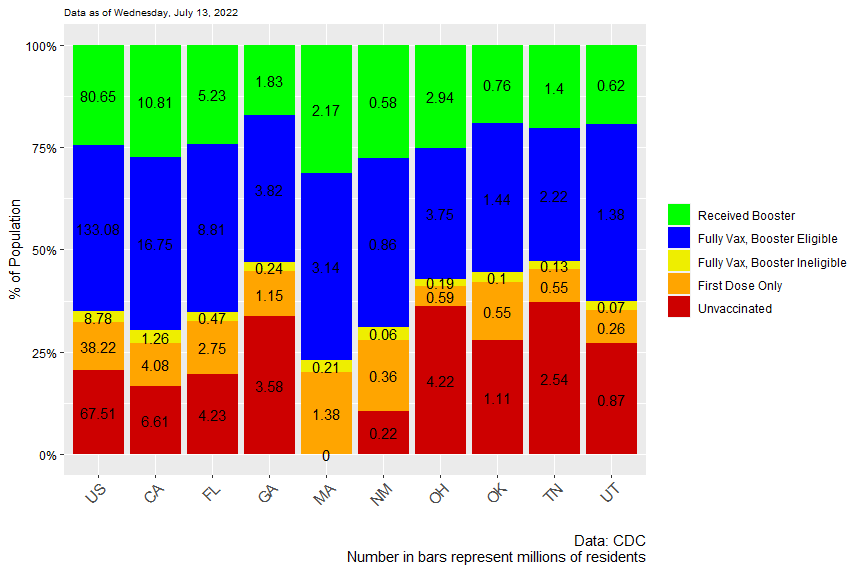
I have some code that downloads COVID vaccination data from the CDCs website, does some manipulation, and eventually produces a bar plot like this
You'll notice the zero at the base of MA's column. The number of unvaccinated people in MA has been getting smaller and smaller and today eventually hit 0. Leaving aside whether you believe there are actually NO people in MA that are unvaccinated, I can believe the number is quite small....
Is there any way that I can tell ggplot to NOT include that 0 data label?
Here's the data
finalBarPlotData <- structure(list(State = structure(c(2L, 3L, 4L, 5L, 6L, 7L, 8L,
9L, 1L, 10L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 1L, 10L, 2L, 3L,
4L, 5L, 6L, 7L, 8L, 9L, 1L, 10L, 2L, 3L, 4L, 5L, 6L, 7L, 8L,
9L, 1L, 10L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 1L, 10L), .Label = c("US",
"CA", "FL", "GA", "MA", "NM", "OH", "OK", "TN", "UT"), class = "factor"),
variable = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L), .Label = c("Boosted",
"Eligible", "Ineligible", "Incomplete", "Unvax"), class = "factor"),
value = c(10811037.5, 5225168.9, 1825544.6, 2168196.8, 581220.4,
2941639.4, 761232.2, 1397638.8, 80651731.4, 624473.9, 16751023.5,
8810766.1, 3818756.4, 3144199.2, 864186.6, 3745298.6, 1439699.8,
2217344.2, 133078622.6, 1382887.1, 1257568, 470455, 236940,
211389, 64755, 192495, 95483, 127987, 8775787, 72985, 4080930,
2745163, 1151722, 1382887, 363931, 592129, 553536, 548320,
38221889, 258139, 6611664, 4226184, 3584460, 0, 222736, 4217538,
1107020, 2537884, 67511493, 867473)), row.names = c(NA, -50L
), class = "data.frame")
and the code for the plot
stackColors <- c(Boosted = "green", Eligible="blue", Ineligible="yellow2",
Incomplete ="orange", Unvax = "red3")
boosterBar<- ggplot(finalBarPlotData, aes(x=State, y=value,
fill=factor(variable,
levels = c("Boosted", "Eligible",
"Ineligible", "Incomplete",
"Unvax"))))
geom_bar(stat="identity", position="fill") #percent stacked
labs(x="", y="% of Population", fill="",
caption="Data: CDC\nNumber in bars represent millions of residents",
#subtitle = paste("Data as of", format(newestDate, "%A, %B %e, %Y")))
scale_y_continuous(labels = function(x) paste0(x*100, "%")) # Multiply by 100 & add %
theme(plot.title = element_text(size = rel(1), face = "bold"),
plot.subtitle = element_text(size = rel(0.7)),
plot.caption = element_text(size = rel(1)),
axis.text.y = element_text(color='black'),
axis.title.y = element_text(color='black'),
axis.text.x = element_text(size=rel(1.25), angle = 45,hjust = 1))
scale_fill_manual(values = stackColors, labels=c("Received Booster","Fully Vax, Booster Eligible","Fully Vax, Booster Ineligible", "First Dose Only", "Unvaccinated"))
geom_text(aes(label=round(value/1000000,digits=2)), position=position_fill(vjust=0.5), color="black")
print(boosterBar)
Thanks in advance....
CodePudding user response:
There's a lot going on in your plot; perhaps the 'easiest' way is to only plot text if it's > 1, e.g.
library(tidyverse)
df <- finalBarPlotData <- structure(list(State = structure(c(2L, 3L, 4L, 5L, 6L, 7L, 8L,
9L, 1L, 10L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 1L, 10L, 2L, 3L,
4L, 5L, 6L, 7L, 8L, 9L, 1L, 10L, 2L, 3L, 4L, 5L, 6L, 7L, 8L,
9L, 1L, 10L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 1L, 10L), .Label = c("US",
"CA", "FL", "GA", "MA", "NM", "OH", "OK", "TN", "UT"), class = "factor"),
variable = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L), .Label = c("Boosted",
"Eligible", "Ineligible", "Incomplete", "Unvax"), class = "factor"),
value = c(10811037.5, 5225168.9, 1825544.6, 2168196.8, 581220.4,
2941639.4, 761232.2, 1397638.8, 80651731.4, 624473.9, 16751023.5,
8810766.1, 3818756.4, 3144199.2, 864186.6, 3745298.6, 1439699.8,
2217344.2, 133078622.6, 1382887.1, 1257568, 470455, 236940,
211389, 64755, 192495, 95483, 127987, 8775787, 72985, 4080930,
2745163, 1151722, 1382887, 363931, 592129, 553536, 548320,
38221889, 258139, 6611664, 4226184, 3584460, 0, 222736, 4217538,
1107020, 2537884, 67511493, 867473)), row.names = c(NA, -50L
), class = "data.frame")
stackColors <- c(Boosted = "green", Eligible="blue", Ineligible="yellow2",
Incomplete ="orange", Unvax = "red3")
boosterBar<- ggplot(finalBarPlotData, aes(x=State, y=value,
fill=factor(variable,
levels = c("Boosted", "Eligible",
"Ineligible", "Incomplete",
"Unvax"))))
geom_bar(stat="identity", position="fill") #percent stacked
labs(x="", y="% of Population", fill="",
caption="Data: CDC\nNumber in bars represent millions of residents")
#subtitle = paste("Data as of", format(newestDate, "%A, %B %e, %Y")))
scale_y_continuous(labels = function(x) paste0(x*100, "%")) # Multiply by 100 & add %
theme(plot.title = element_text(size = rel(1), face = "bold"),
plot.subtitle = element_text(size = rel(0.7)),
plot.caption = element_text(size = rel(1)),
axis.text.y = element_text(color='black'),
axis.title.y = element_text(color='black'),
axis.text.x = element_text(size=rel(1.25), angle = 45,hjust = 1))
scale_fill_manual(values = stackColors, labels=c("Received Booster","Fully Vax, Booster Eligible","Fully Vax, Booster Ineligible", "First Dose Only", "Unvaccinated"))
geom_text(aes(label=ifelse(value > 1, round(value/1000000,digits=2), "")), position=position_fill(vjust=0.5), color="black")
print(boosterBar)
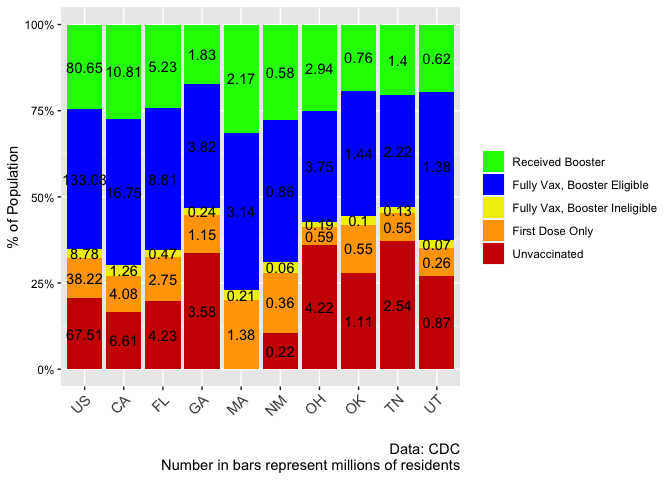
Created on 2022-07-21 by the reprex package (v2.0.1)