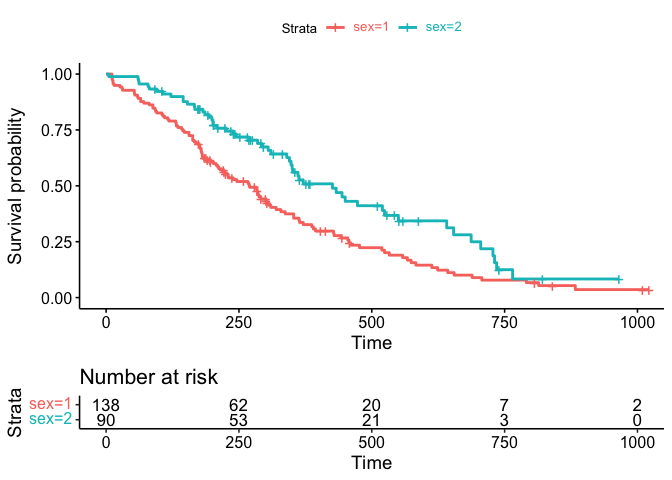
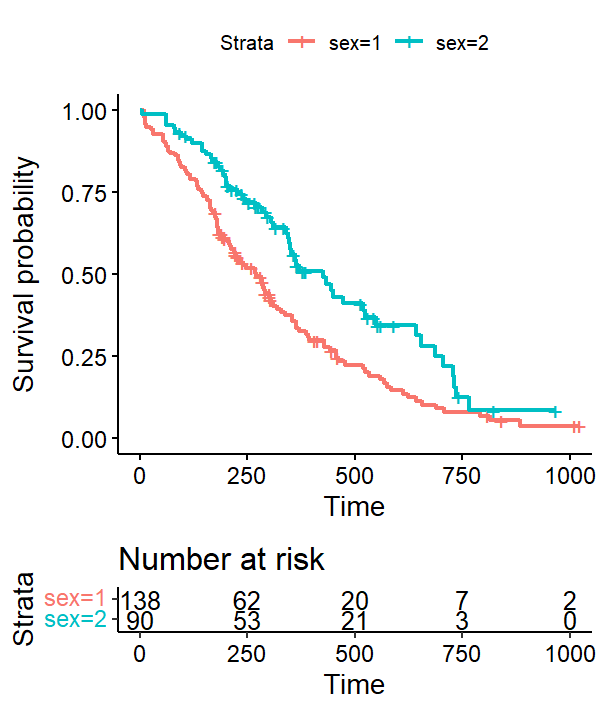
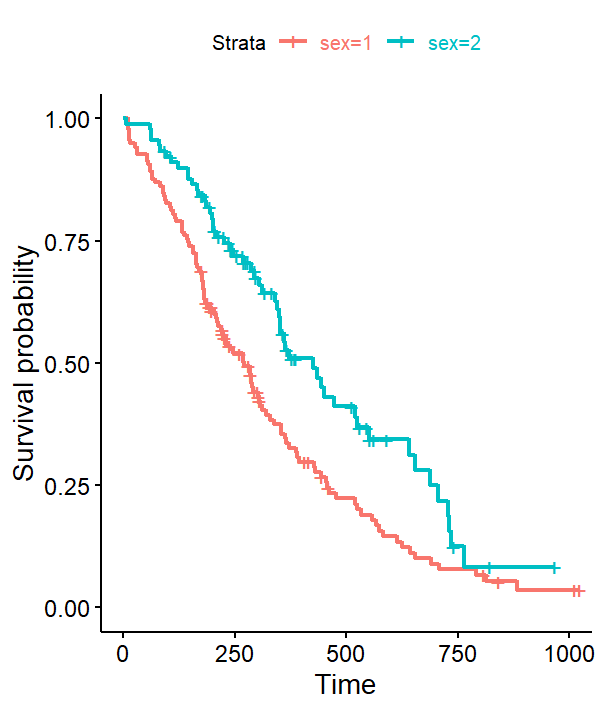
What I would really like to do is change the color of the text on the legend to match the color of the lines on the graph.
Where I am stuck is now integrating this with the original ggsurvplot risk table underneath, as in the first graph.
My pretty basic reaction was to simply replace the ggsurvplot$plot with the new ggplot object created after extracting the original plot to a grob and then back to a ggplot...
g1$plot <- plot1
However this does not work...
Error in
[.data.frame(g$data[1], "colour") : undefined columns selected
Most likely due to a loss of the underlying data in the process above, storing only a 2x2 table...
plot1$data
x y
1 0 0
2 1 1
Compared to the original fitgraph$plot$data which yields the entire data set (hundreds of rows, ongoing survival proportions) which presumably feeds the risk table. The other dumb strategy of plot1 fitgraph$table doesn't work either.
There must be a better strategy - any ideas? Thanks in advance!
CodePudding user response:
While I appreciate your effort the ggtext package offers an easy option to achieve your desired result. Besides making it easier to set the legend text colors the final result could simply assigned back to the plot element of the ggurvplot object:
library(survival)
library(survminer)
library(ggtext)
fit <- survfit(Surv(time, status) ~ sex, data = lung)
fitgraph <- ggsurvplot(fit, risk.table = TRUE, risk.table.y.text.col = TRUE)
cols <- scales::hue_pal()(2)
labels <- function(x, cols) {
glue::glue("<span style = 'color: {cols}'>{x}</span>")
}
fitgraph$plot <- fitgraph$plot
scale_color_discrete(labels = ~labels(.x, cols))
theme(legend.text = element_markdown())
fitgraph