I am trying to create two figures with squared subfigures for an article. My problem is that when I do it for two subfigures I get one size and when I do it for three I get a different one, and I would like both to have the same overall size. Moreover, I would like to have all subfigures touching each other as in the examples below.
I am sharing the codes and the outputs for both cases.
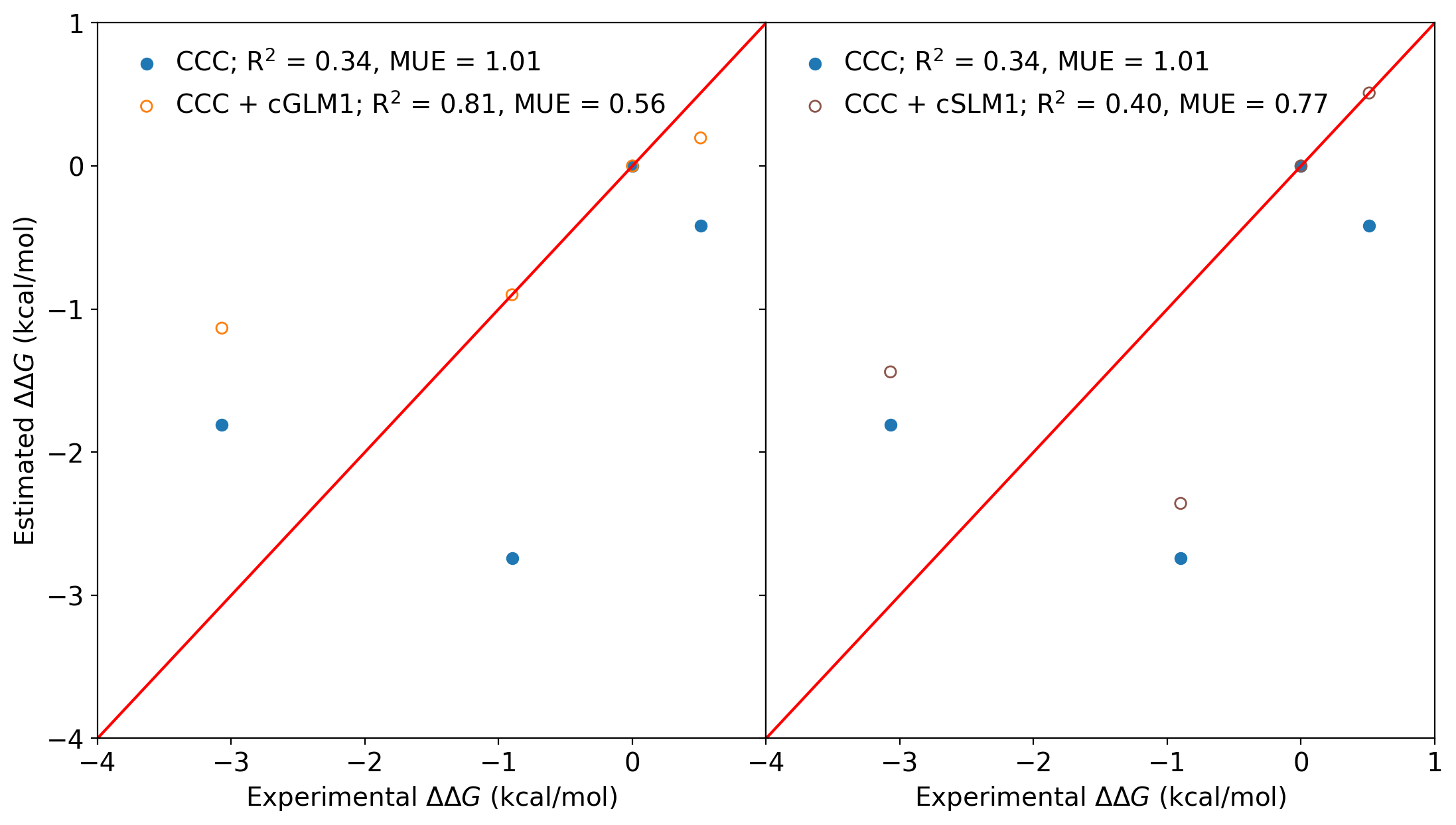
Two subfigures:
fig, ax = plt.subplots(1,2, figsize=(13,7), sharey=True, facecolor='w')
fig.subplots_adjust(wspace=0, hspace=0)
ax[0].scatter(exp_list, yrel_slack_list, label='CCC; R$^2$ = 0.34, MUE = 1.01')
ax[0].scatter(exp_list, yrel_glm1_list, facecolors='none', edgecolors='#ff7f0e', label='CCC cGLM1; R$^2$ = 0.81, MUE = 0.56')
ax[0].plot(np.linspace(-4,4), np.linspace(-4,4), color='red')
ax[1].scatter(exp_list, yrel_slack_list, label='CCC; R$^2$ = 0.34, MUE = 1.01')
ax[1].scatter(exp_list, yrel_slm1_list, facecolors='none', edgecolors='#8c564b', label='CCC cSLM1; R$^2$ = 0.40, MUE = 0.77')
ax[1].plot(np.linspace(-4,4), np.linspace(-4,4), color='red')
ax[0].set_ylabel(r'Estimated $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax[0].set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax[0].legend(loc=0, frameon=False, fontsize=14, handletextpad=0.1)
ax[0].set_ylim([-4,1])
ax[0].set_xlim([-4,1])
ax[0].set_xticks([-4, -3, -2, -1, 0])
ax[0].tick_params(axis = 'both', which = 'major', labelsize=14)
ax[1].set_xlim([-4,1])
ax[1].legend(loc=0, frameon=False, fontsize=14, handletextpad=0.1)
ax[1].set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax[1].tick_params(axis = 'both', which = 'major', labelsize=14)
plt.savefig('inclusion_1h1q_correlation.eps', format='eps', bbox_inches='tight')
plt.show()
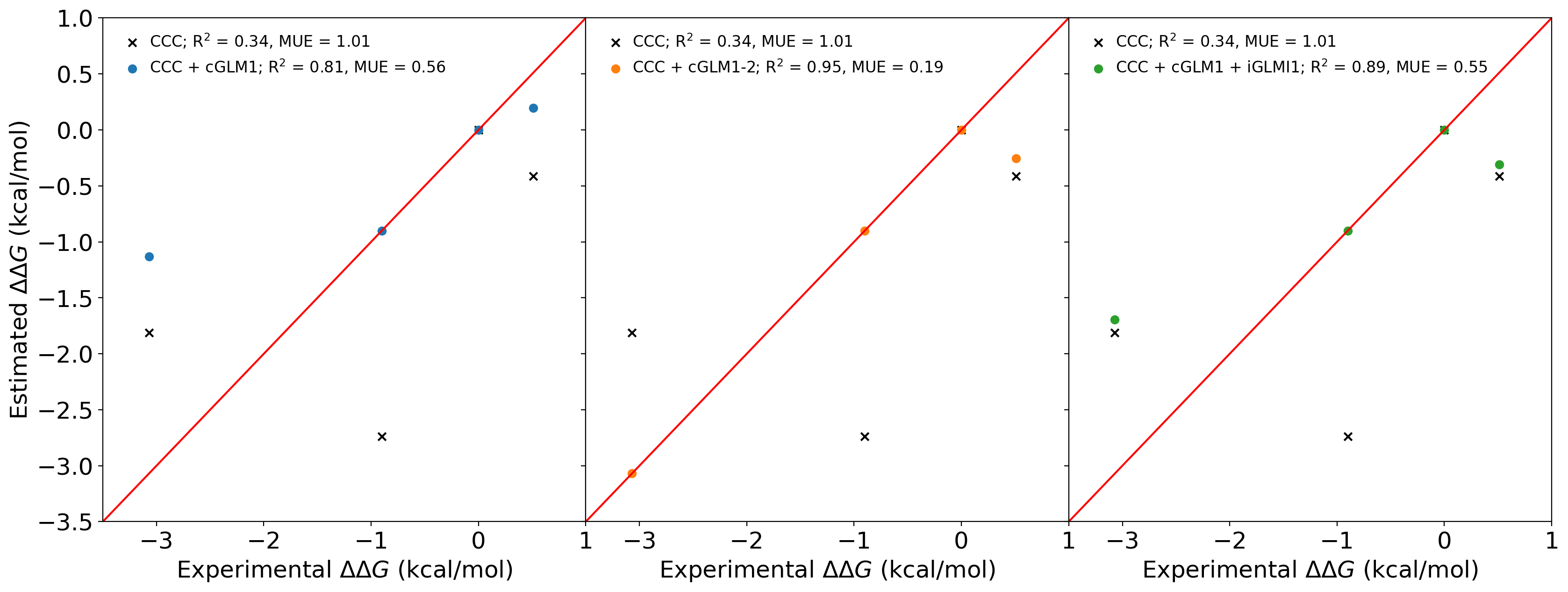
Three subfigures:
fig, ax = plt.subplots(1,3, figsize=(20,7), sharey=True, facecolor='w')
fig.subplots_adjust(wspace=0, hspace=0)
ax[0].scatter(exp_list, yrel_slack_list, marker='x', color='k', label='CCC; R$^2$ = 0.34, MUE = 1.01')
ax[0].scatter(exp_list, yrel_c1oiu1h1q_list, color='#1f77b4', label='CCC cGLM1; R$^2$ = %.2f, MUE = %.2f'%(corr_node_c1oiu1h1q[0]**2,corr_node_c1oiu1h1q[1]))
ax[0].plot(np.linspace(-4,1), np.linspace(-4,1), color='red')
ax[1].scatter(exp_list, yrel_slack_list, marker='x', color='k', label='CCC; R$^2$ = 0.34, MUE = 1.01')
ax[1].scatter(exp_list, yrel_c1oiu1h1q_c1oiu1h1s_list, color='#ff7f0e', label='CCC cGLM1-2; R$^2$ = %.2f, MUE = %.2f'%(corr_node_c1oiu1h1q_c1oiu1h1s[0]**2,corr_node_c1oiu1h1q_c1oiu1h1s[1]))
ax[1].plot(np.linspace(-4,1), np.linspace(-4,1), color='red')
ax[2].scatter(exp_list, yrel_slack_list, marker='x', color='k', label='CCC; R$^2$ = 0.34, MUE = 1.01')
ax[2].scatter(exp_list, yrel_c1h1q1oiu_i1oiu_list, color='#2ca02c', label='CCC cGLM1 iGLMI1; R$^2$ = %.2f, MUE = %.2f'%(corr_node_1oiu[0]**2,corr_node_1oiu[1]))
ax[2].plot(np.linspace(-4,1), np.linspace(-4,1), color='red')
ax[0].set_ylabel(r'Estimated $\Delta \Delta G$ (kcal/mol)', fontsize=18)
ax[0].set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=18)
ax[0].legend(loc=0, frameon=False, fontsize=12, handletextpad=0.1)
ax[0].set_ylim([-3.5,1])
ax[0].set_xlim([-3.5,1])
ax[0].tick_params(axis = 'both', which = 'major', labelsize=18)
ax[1].set_xlim([-3.5,1])
ax[1].legend(loc=0, frameon=False, fontsize=12, handletextpad=0.1)
ax[1].set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=18)
ax[1].tick_params(axis = 'both', which = 'major', labelsize=18)
ax[2].set_xlim([-3.5,1])
ax[2].legend(loc=0, frameon=False, fontsize=12, handletextpad=0.1)
ax[2].set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=18)
ax[2].tick_params(axis = 'both', which = 'major', labelsize=18)
plt.savefig('main_result_cdk2.eps', format='eps', bbox_inches='tight')
plt.show()
CodePudding user response:
You can use gridspec to align subplots ratios for a given overall figure size incl. your required y-axis touching.
I haven't adapted all your various individual plot settings, but tried to show how gridspec can be used for your general case and kept the respective parts like e.g. the axis ticks (to not overlap especially at the shared y-axis).
from matplotlib import gridspec
### 2 subplots
fig = plt.figure(figsize=(20,7), facecolor='w')
fig.subplots_adjust(wspace=0, hspace=0)
gs = gridspec.GridSpec(1, 2, width_ratios=[1,1])
ax1 = fig.add_subplot(gs[0,:1])
ax1.set_title('ax1')
ax1.set_ylim([-4,1])
ax1.set_xlim([-4,1])
ax1.set_xticks([-4, -3, -2, -1, 0])
xx, locs = plt.yticks()
ll = ['%.1f' % a for a in xx]
ax1.set_yticks(xx, ll)
ax1.tick_params(axis = 'both', which = 'major', labelsize=14)
ax2 = fig.add_subplot(gs[0,1:])
ax2.set_title('ax2')
ax2.get_yaxis().set_visible(False)
ax2.set_xlim([-4,1])
ax2.tick_params(axis = 'both', which = 'major', labelsize=14)
ax1.set_ylabel(r'Estimated $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax1.set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax2.set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
plt.show()
### 3 subplots
fig = plt.figure(figsize=(20,7), facecolor='w')
fig.subplots_adjust(wspace=0, hspace=0)
gs = gridspec.GridSpec(1, 3, width_ratios=[1,1,1])
ax1 = fig.add_subplot(gs[0,:1])
ax1.set_title('ax1')
ax1.set_ylim([-3.5,1])
ax1.set_xlim([-3.5,1])
ax1.tick_params(axis = 'both', which = 'major', labelsize=14)
ax2 = fig.add_subplot(gs[0,1])
ax2.set_title('ax2')
ax2.get_yaxis().set_visible(False)
ax2.set_xlim([-3.5,1])
ax2.tick_params(axis = 'both', which = 'major', labelsize=14)
ax3 = fig.add_subplot(gs[0,2:])
ax3.set_title('ax3')
ax3.get_yaxis().set_visible(False)
ax3.set_xlim([-3.5,1])
ax3.tick_params(axis = 'both', which = 'major', labelsize=14)
ax1.set_ylabel(r'Estimated $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax1.set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax2.set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
ax3.set_xlabel('Experimental $\Delta \Delta G$ (kcal/mol)', fontsize=14)
plt.show()
Notes:
- The inner y-axes need the
set_visible(False) - With
figsize=(20,7)on top you can adapt the overal plot size- You may align that if you need more squared subplots and accept different heights for that at a given overall width.
- If you want to keep the ticks on the connected y-axes but only want to remove the label you can replace
ax2.get_yaxis().set_visible(False)byax2.tick_params(axis='y', label1On=False) - Items to fine tune the optical width on the left y-axis - adapt those acc. your preference.
- I notized you had different font sizes and aligned them.
- At the 2 subplots y-axis floats with 1 decimal point is included (as given by the 3 subplots y-axis), you can remove that part again easily if it's not intended.
Here's a screenshot incl. some jupyter parts to show that both overall plots have the same size: