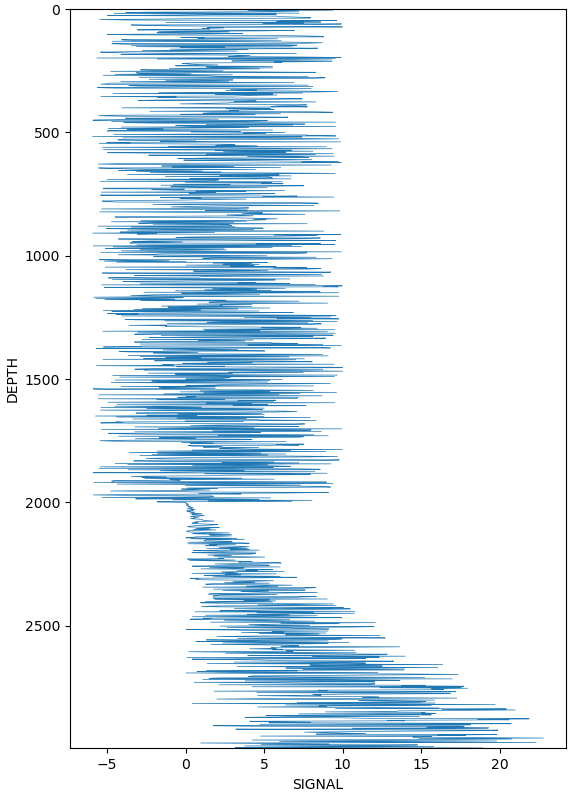
This is an example of a larger data with many dataframes similar to this one below (df_final):
df1 = pd.DataFrame({"DEPTH (m)":np.arange(0, 2000, 2),
"SIGNAL":np.random.uniform(low=-6, high=10, size=(1000,))})
df2 = pd.DataFrame({"DEPTH (m)":np.arange(2000, 3000, 2),
"SIGNAL":np.random.uniform(low=0, high=5, size=(500,))})
for i, row in df2.iterrows():
df2.loc[i, "SIGNAL"] = row["SIGNAL"] * (i / 100)
df_final = pd.concat([df1, df2])
You can see that this signal has two patterns (one "constant" and other increasing):
plt.figure()
plt.plot(df_final["SIGNAL"], df_final["DEPTH (m)"], linewidth=0.5)
plt.ylim(df_final["DEPTH (m)"].max(), df_final["DEPTH (m)"].min())
plt.xlabel("SIGNAL")
plt.ylabel("DEPTH")
Is there a way I can automatically create a flag/cluster to split this signal? In this example I would have one cluster before depth 2000 and other after it.
Another problem is that, in my project, I will have other dataframes with more than two signal patterns and couldn't set it manually for each dataframe as there are many.
CodePudding user response:
To cluster the time series into different patterns in this case, you can use clustering algorithms such as KMeans or DBSCAN. there is an example of how to do it with KMeans:
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
# Scale the data
scaler = StandardScaler()
df_final_scaled = scaler.fit_transform(df_final[["SIGNAL"]])
# Fit KMeans
kmeans = KMeans(n_clusters=2)
kmeans.fit(df_final_scaled)
# Predict the clusters
df_final_clusters = kmeans.predict(df_final_scaled)
# Visualize the results
plt.figure()
plt.scatter(df_final["SIGNAL"], df_final["DEPTH (m)"], c=df_final_clusters, cmap="viridis")
plt.xlabel("SIGNAL")
plt.ylabel("DEPTH")
plt.show()
CodePudding user response:
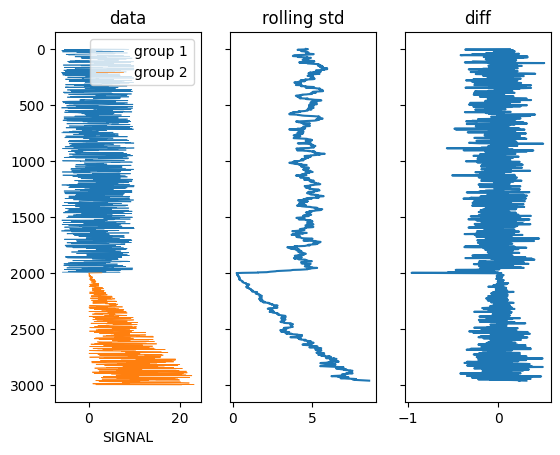
One possibility using a rolling standard deviation:
s1 = df_final.loc[::-1, 'SIGNAL'].rolling(20).std()[::-1]
s2 = s1.diff()
N = 2 # number of groups
m = s2.lt(s2.quantile((N-1)/len(df_final)))
groups = (m&~m.shift(fill_value=False)).cumsum()
f, (ax, ax1, ax2) = plt.subplots(ncols=3, sharey=True)
for k, g in df_final.groupby(groups):
g.plot(x='SIGNAL', y='DEPTH (m)', ax=ax, lw=0.5, label=f'group {k 1}')
ax1.plot(s1, df_final['DEPTH (m)'])
ax2.plot(s2, df_final['DEPTH (m)'])
ax.invert_yaxis()
ax.set_title('data')
ax1.set_title('rolling std')
ax2.set_title('diff')
Output: