I am trying to plot a qq function of a custom CDF function, of the form f(x)= p*e^-x (1-p)*e^-5x, p being between 0 and 1, and x greater than 0. Data has values 0<x<500,000
In doing this I have stumbled across the uniroot function which looks to solve inverse functions. I am running into a few errors however, and would greatly appreciate help on this.
My goal here is writing a function along the lines of R's base functions qnorm qexp etc. but with qcustom.
# Error 1: (Error in f(x) : argument "p" is missing, with no default)
inverse <- function(f, lower=0.01, upper=500000) {
function(y) {
uniroot(function(x) f(x) - y, lower=lower, upper=upper)[1]
}
}
qcustom <- inverse(function(x, p, a, b) {
a1 <- p*exp(-a*x)
a2 <- (1 - p)*exp(-b*x)
result <- 1 - a1 - a2
return(result)
})
qcustom(0.5) ##when i add in further parameters, e.g p = 0.5 it says it's an unused parameter##
CodePudding user response:
Let's start at the beginning:
custom_cdf <- function(x, p, a, b) {
p*exp(-a*x) (1-p)*exp(-b*x)
}
This has parameters c(p, a, b) and let us try to find a pairing of c(a,b) that
makes sense for the range(x) = c(0, 500e3) as you pointed out that you want.
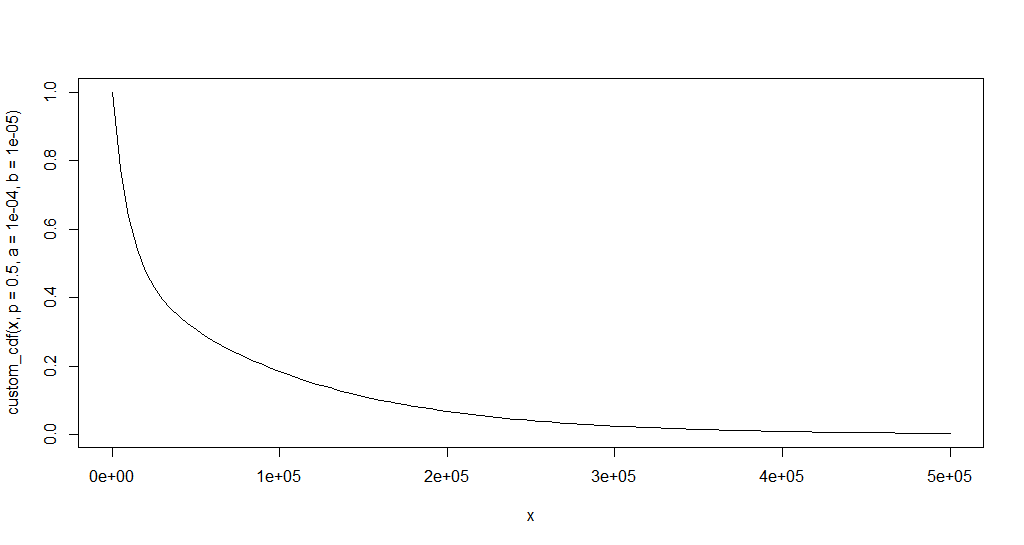
plot.new()
curve(custom_cdf(x, p = 0.5, a = 1e-4, b = 1e-5), xlim = c(0, 500e3))
A reasonable set of values could be:
a <- 1e-4
b <- 1e-5
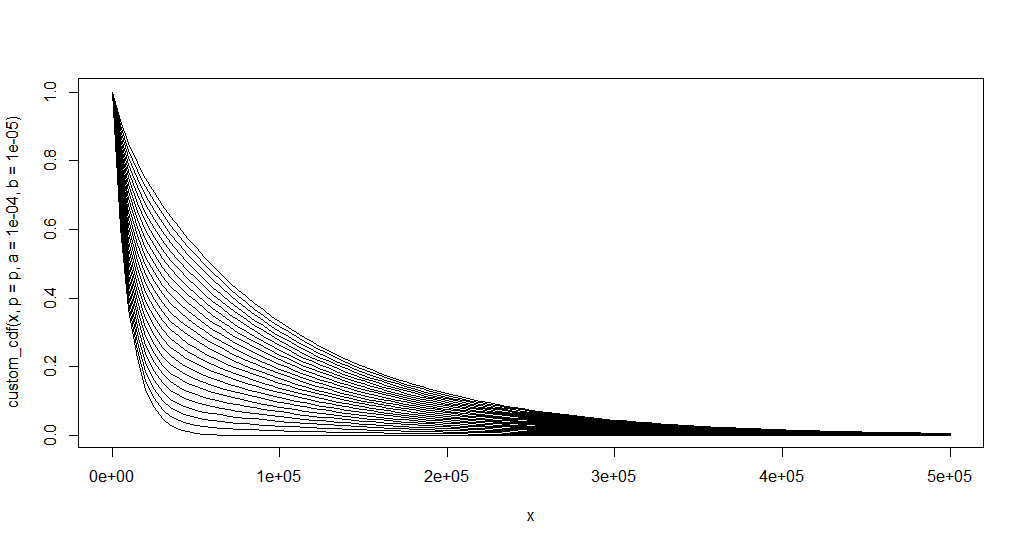
Let us try for multiple values of p:
first <- TRUE
plot.new()
for (p in seq.default(0.1, 1, length.out = 25)) {
curve(custom_cdf(x, p = p, a = 1e-4, b = 1e-5), xlim = c(0, 500e3), add = !first)
first <- FALSE
}
rm('p')
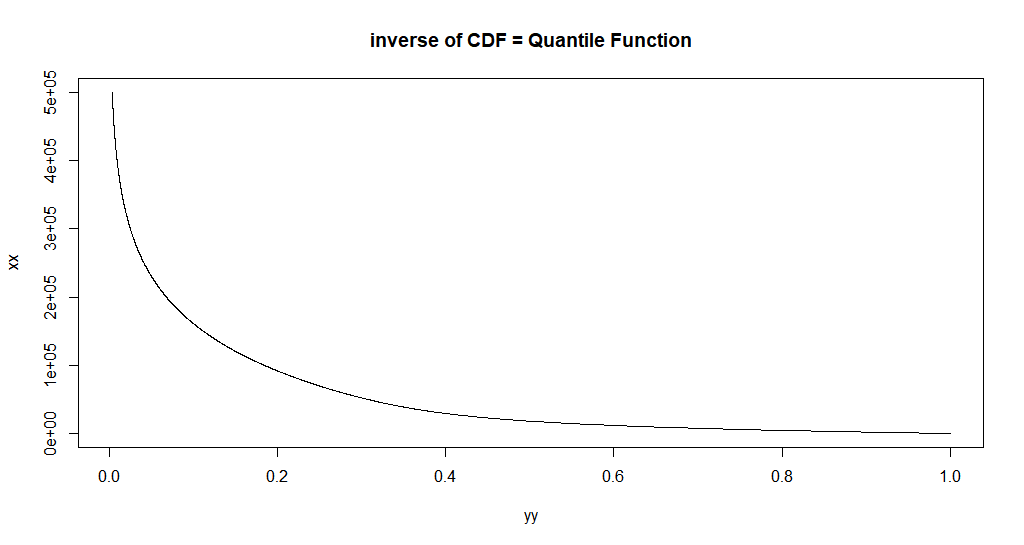
To just the inverse of the CDF, we could just:
xx <- seq.default(0, 500e3, length.out = 2500)
# xx <- seq.default(0, 500e3, by = 10)
# xx <- seq.default(0, 500e3, by = 1)
yy <- custom_cdf(xx, p = 0.5, a = a, b = b)
plot.new()
plot(yy, xx, main = "inverse of CDF = Quantile Function",type = 'l')
I.e. we can just reverse the plotting. So now, this is a picture of the inverse function.
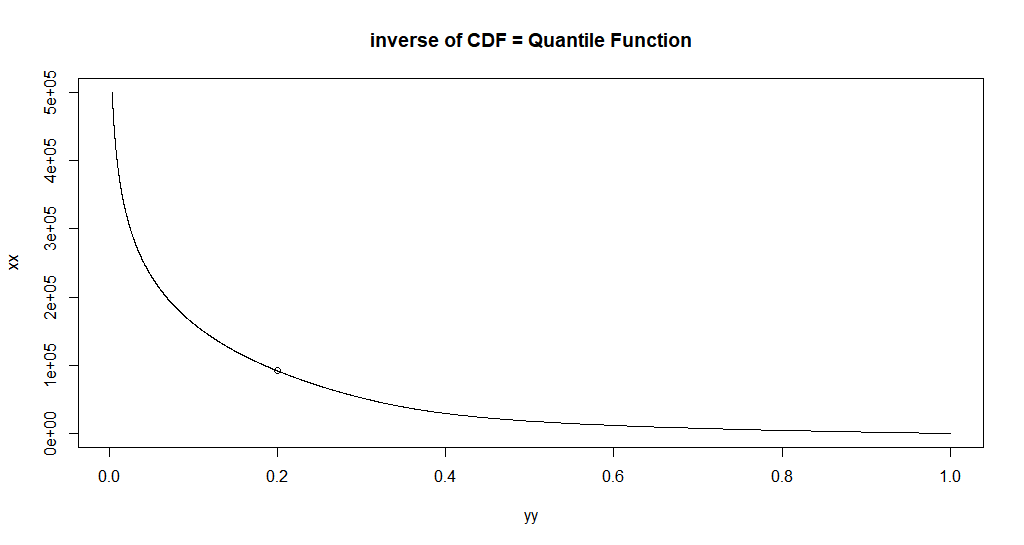
Let's use uniroot to get one inverse value.
uniroot(
function(x) custom_cdf(x, p = 0.5, a = a, b = b) - 0.2,
interval = c(0, 100e50),
extendInt = "yes"
) -> found_20pct_point
#'
#' Let's plot that point:
#'
points(custom_cdf(found_20pct_point[[1]], p = 0.5, a = a, b = b), found_20pct_point[[1]])
Alright, so now we understand everything and we can do what you tried to do:
custom_quantile <- function(p, a, b) {
particular_cdf <- function(x) custom_cdf(x, p, a, b)
function(prob) {
uniroot(
function(x) particular_cdf(x) - prob,
interval = c(0, 100e50),
extendInt = "yes"
)$root
}
}
custom_quantile_specific <-
custom_quantile(p = 0.5, a = a, b = b)
custom_quantile_specific(0.2)
And this works as it yields something close to the desired value.
But regular quantile functions in R does depend on the parameters of the distribution, so this is good enough:
custom_quantile <- function(prob, p, a, b) {
particular_cdf <- function(x) custom_cdf(x, p, a, b)
uniroot(
function(x) particular_cdf(x) - prob,
interval = c(0, 100e50),
extendInt = "yes"
)$root
}
custom_quantile(0.2, p = 0.5, a = a, b = b)