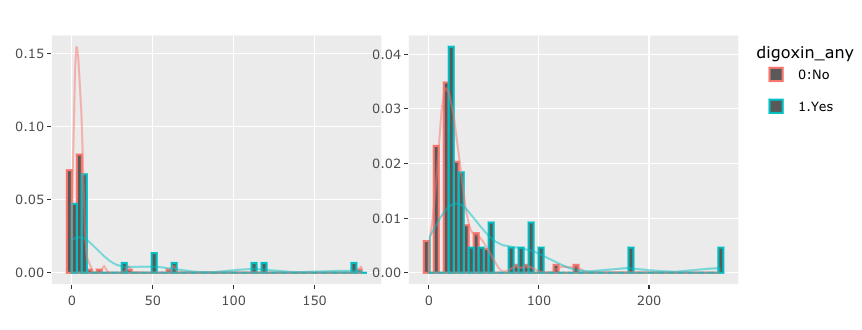
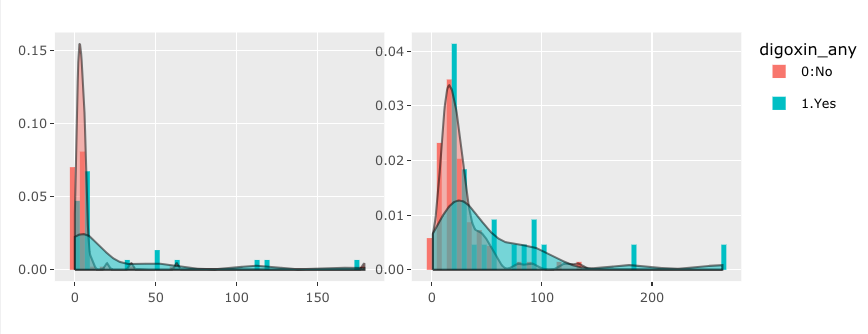
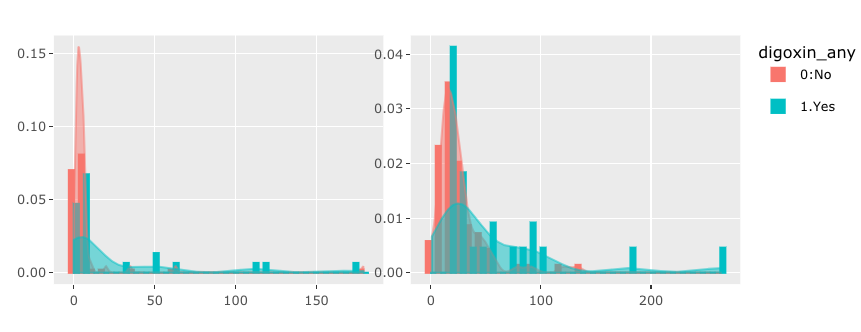
I would like to make an interactive graphs based on user input. However I'm struggle to make more than one graphs using R plotly. Suppose I have following data and codes,
dput(norwd5)
structure(list(LENGTH_OF_STAY = c(57L, 28L, 15L, 28L, 14L, 49L,
15L, 22L, 17L, 81L, 34L, 24L, 31L, 38L, 33L, 22L, 21L, 49L, 188L,
21L, 21L, 36L, 24L, 23L, 48L, 54L, 42L, 62L, 13L, 139L, 29L,
49L, 15L, 7L, 43L, 28L, 31L, 22L, 23L, 26L, 33L, 30L, 127L, 22L,
22L, 15L, 28L, 26L, 15L, 31L, 22L, 89L, 28L, 60L, 54L, 37L, 20L,
135L, 155L, 51L, 15L, 8L, 38L, 16L, 16L, 22L, 30L, 14L, 16L,
18L, 14L, 272L, 25L, 22L, 18L, 21L, 188L, 264L, 34L, 34L, 136L,
23L, 142L, 25L, 32L, 58L, 163L, 16L, 35L, 23L, 50L, 71L, 10L,
19L, 22L, 24L, 45L, 29L, 15L, 82L), PRE_OPERATIVE_LOS = c(2L,
2L, 3L, 1L, 3L, 6L, 3L, 7L, 2L, 2L, 11L, 2L, 6L, 3L, 6L, 3L,
5L, 3L, 179L, 2L, 5L, 3L, 4L, 2L, 5L, 6L, 2L, 4L, 2L, 6L, 3L,
2L, 2L, 6L, 6L, 1L, 4L, 5L, 6L, 5L, 0L, 4L, 6L, 2L, 4L, 4L, 7L,
4L, 4L, 6L, 2L, 4L, 3L, 3L, 2L, 6L, 4L, 110L, 63L, 6L, 4L, 7L,
5L, 1L, 6L, 1L, 4L, 2L, 6L, 3L, 2L, 8L, 2L, 2L, 4L, 3L, 6L, 171L,
5L, 4L, 116L, 6L, 47L, 3L, 7L, 3L, 60L, 1L, 3L, 20L, 31L, 49L,
9L, 8L, 3L, 4L, 35L, 7L, 4L, 9L), POST_OPERATIVE_LOS = c(55L,
26L, 12L, 27L, 11L, 43L, 12L, 15L, 15L, 79L, 23L, 22L, 25L, 35L,
27L, 19L, 16L, 46L, 9L, 19L, 16L, 33L, 20L, 21L, 43L, 48L, 40L,
58L, 11L, 133L, 26L, 47L, 13L, 1L, 37L, 27L, 27L, 17L, 17L, 21L,
33L, 26L, 121L, 20L, 18L, 11L, 21L, 22L, 11L, 25L, 20L, 85L,
25L, 57L, 52L, 31L, 16L, 25L, 92L, 45L, 11L, 1L, 33L, 15L, 10L,
21L, 26L, 12L, 10L, 15L, 12L, 264L, 23L, 20L, 14L, 18L, 182L,
93L, 29L, 30L, 20L, 17L, 95L, 22L, 25L, 55L, 103L, 15L, 32L,
3L, 19L, 22L, 1L, 11L, 19L, 20L, 10L, 22L, 11L, 73L), digoxin_any = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L,
2L, 1L, 2L), .Label = c("0:No", "1.Yes"), class = "factor")), row.names = c(NA,
-100L), class = c("data.table", "data.frame"), .internal.selfref = <pointer: 0x0000012f36b61ef0>)
num <- c('PRE_OPERATIVE_LOS','POST_OPERATIVE_LOS')
plist <- scan(text=num,what = "",quiet = T)
groups <- 'digoxin_any'
bygrp <- scan(text=groups,what="",quiet=T)
norwd5[, (bygrp) := lapply(.SD, as.factor), .SDcols = bygrp]
plotList = list()
for(i in length(plist)){
gplot <- ggplot(norwd5,aes_string(x=plist[i],group=bygrp,color=bygrp)) geom_histogram(aes(y=..density..),position = "dodge") geom_density(alpha=.5) theme(legend.position = "left")
plotList[[i]] <- plotly_build(gplot)
}
for(i in length(plist)){
print(plotList[[i]])
}
The goal is to show both graphs for PRE_OPERATIVE_LOS and POST_OPERATIVE_LOS. However, the codes above only show histogram for POST_OPERATIVE_LOS.
I checked maybe subplot is the way to go but how to make subplot work in a loop? Any hints? Thanks!
CodePudding user response:
There is an error in your first loop and calling each subplot won't make both appear at the same time.
First-- the issue with your first for call- when you wrote
for(i in length(plist))
You wrote for i in 2 or i == 2, meaning that you never looped. If you modify it to a range of values, now it's written: for i in 1 to 2.
for(i in 1:length(plist))
So you're aware, if you had written for(i in plist) it would have done both loops, but instead of a value, i would be the strings.
Okay, so now there are two graphs. From the plotly library, you can use the function subplot. You will want to turn the legend off for one of them, though.
subplot(plotList[[1]],
style(plotList[[2]], showlegend = FALSE))
If you wanted the outline color, that's more than okay! However, if you wanted to bars to be filled, you need to assign fill instead of color.
If you change color = bygrp to fill = bygrp, this is how this would change:
If you leave the color assignment and add fill = bygrp (so you have both), this is how this would change: