Still a novice with R.
Background
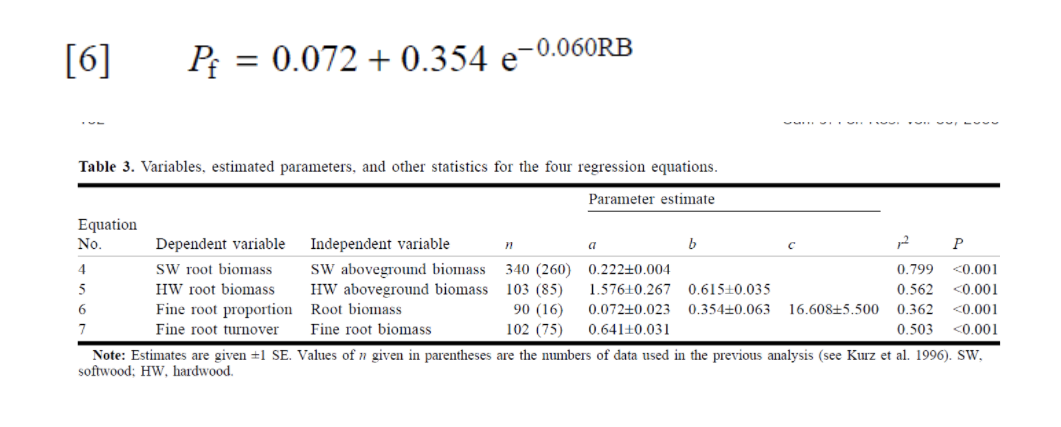
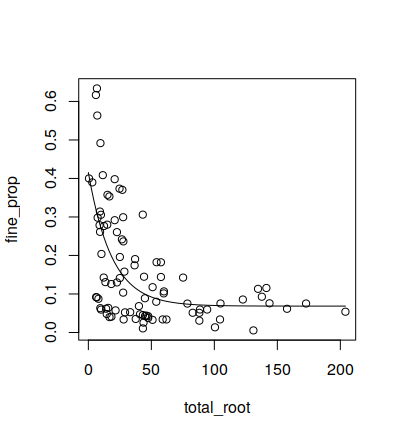
I have the data from Li et al. 2003 paper "Belowground biomass dynamics in the Carbon Budget Model of the Canadian Forest Sector".(
CodePudding user response:
We can use nls as follows.
fit = nls(fine_prop ~ m * (1 - b * exp(c*total_root)),
data = fr,
start = list(m=0.1, b = 2, c=-0.05))
coefficients(fit)
# m b c
# 0.06851410 -5.06265496 -0.05369425
plot(fr)
x = seq(0, max(fr$total_root), length.out=100)
lines(x, predict(fit, newdata = data.frame(total_root = x)))
Equivalently, we can also put the equation in the same format as in the published article, rather than the form in OP. The we have:
fit = nls(fine_prop ~ a b * exp(c*total_root),
data = fr,
start = list(a=1, b = 1, c=-0.1))
coefficients(fit)
# a b c
# 0.0685124 0.3468601 -0.0536925
The data:
fr = structure(list(total_root = c(28, 47.5, 104.5, 62, 59.8304, 50.9335,
40.1412, 43.3121, 131.0057, 24.74, 137.71, 25.004, 88.1, 88.1,
57.6, 57.6, 54.16, 82.82, 88.6, 134.68, 20.92, 25.004, 141.31,
143.8, 204.04, 104.88, 172.8, 122.62, 157.7, 18.184, 13.538,
10.4, 27, 36.7, 44.2, 53.9, 78.6, 47.5, 6.6, 10.1, 14.6, 16.8,
18.3, 8, 9.5, 6.2, 14.1, 15.8, 21.6, 29.1, 33.2, 41, 45, 46,
59, 37.6, 9.3, 15, 22.7, 43.2, 37.1, 51, 28.6, 60, 75.2, 12.2,
43.8, 43.3, 6.8, 6, 7.1, 8.9, 94.3, 100.5, 44.9, 9.98, 9.17,
7.38, 20.9, 9.6, 11.48, 22.68, 27.63, 27.63, 15.19, 27.78, 26.63,
12.44, 16.7, 0.5, 3.13), fine_prop = c(0.033796, 0.037486, 0.033818,
0.033774, 0.10147, 0.032788, 0.068508, 0.045253, 0.005412, 0.373484,
0.092876, 0.196049, 0.030647, 0.051078, 0.144097, 0.182292, 0.182422,
0.051195, 0.059594, 0.113306, 0.291587, 0.141337, 0.115562, 0.0758,
0.053911, 0.075324, 0.075231, 0.08563, 0.061509, 0.125825, 0.130743,
0.203846, 0.37037, 0.174387, 0.144796, 0.079777, 0.075064, 0.042316,
0.090909, 0.059406, 0.047945, 0.039881, 0.040984, 0.0875, 0.063158,
0.091935, 0.061702, 0.063924, 0.057407, 0.052234, 0.052711, 0.047317,
0.042667, 0.043913, 0.033898, 0.035106, 0.26129, 0.28, 0.129956,
0.306019, 0.190566, 0.117647, 0.158042, 0.106667, 0.142553, 0.142623,
0.025571, 0.010624, 0.633824, 0.616667, 0.56338, 0.278652, 0.059597,
0.013433, 0.088864, 0.305611, 0.314068, 0.298103, 0.398086, 0.491667,
0.408537, 0.260582, 0.299312, 0.103511, 0.357472, 0.236501, 0.241833,
0.276527, 0.353293, 0.4, 0.389776)), row.names = c(NA, -91L), class = "data.frame")