I can't figure out to plot my data using geom_line and geom_point so that gghighlight works without needing the variables to be in the global environment
library(ggplot2)
library(gghighlight)
Create some data
x <- rep(1:5, 4)
y <- c(rep(1:5, 1) rnorm(5)/5, rep(1:5, 1) rnorm(5)/5 1, rep(6:10, 1) rnorm(5)/5, rep(6:10, 1) rnorm(5)/5 1)
treatment <- gl(2, 10, 20, labels=letters[1:2])
replicate <- gl(2, 5, 20, labels=letters[25:26])
d <- data.frame(x=x, y=y, treatment=treatment, replicate=replicate)
d
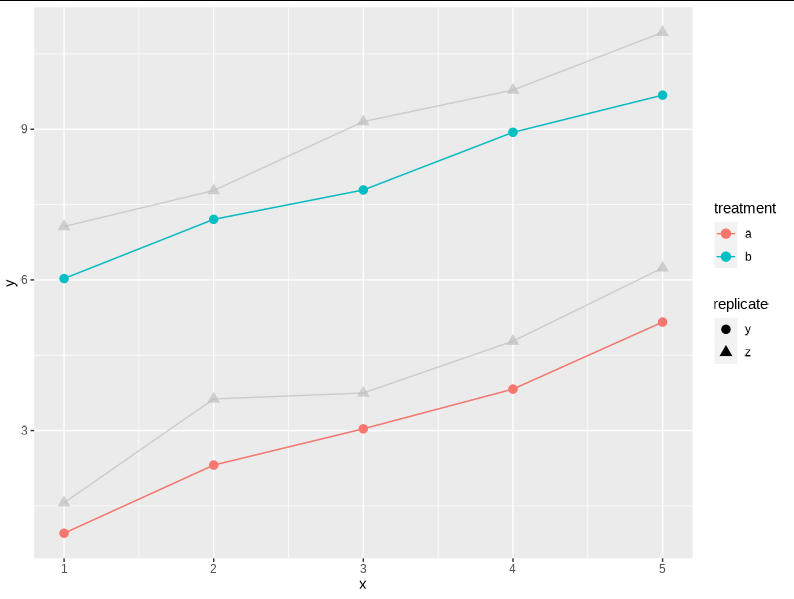
Plot v1 is as desired
v1 <- ggplot(d, aes(x=x, y=y, colour=treatment, shape = replicate, group=interaction(treatment, replicate)))
geom_point(size = 3) geom_line()
v1
gghighlight works here as required but only when the variables are in the global environment
v1 gghighlight(replicate == "y")
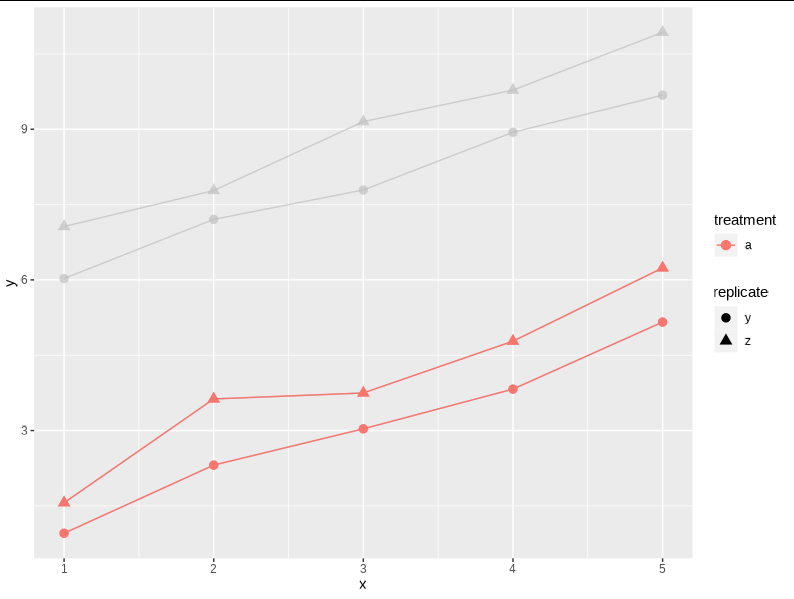
v1 gghighlight(treatment == "a")
rm(replicate, treatment,x,y)
v1 gghighlight(replicate == "y")
v1 gghighlight(treatment == "a")
if the global variables are rmeoved I get this error message
Error in `f()`:
! Aesthetics must be valid data columns. Problematic aesthetic(s): shape = replicate.
Did you mistype the name of a data column or forget to add after_stat()?
Run `rlang::last_error()` to see where the error occurred.
Warning messages:
1: Tried to calculate with group_by(), but the calculation failed.
Falling back to ungrouped filter operation...
2: Using `across()` in `filter()` is deprecated, use `if_any()` or `if_all()`.
3: Tried to calculate with group_by(), but the calculation failed.
Falling back to ungrouped filter operation...
4: Using `across()` in `filter()` is deprecated, use `if_any()` or `if_all()`. `
Try alternative ways of generating the plot
v2 <- ggplot(d, aes(x=x, y=y, colour=treatment, shape = replicate))
geom_point(size=3) geom_line()
v2
Highlighting does not work
v2 gghighlight(replicate == "y")
v2 gghighlight(treatment == "a")
Third version of plot
v3 <- ggplot(d)
geom_point(aes(x=x, y=y, colour=treatment, shape = replicate), size = 3)
geom_line(aes(x=x, y=y, colour=treatment, shape = replicate))
v3
Highlight occurs but goes wrong
v3 gghighlight(replicate == "y")
v3 gghighlight(treatment == "a")
CodePudding user response:
I think the problem lies with you mapping the shape aesthetic in the main ggplot call, instead of specifying it within the geom_point call. The gghighlight function creates a new data frame for each layer with the required aesthetic values, and this process doesn't work properly if there is unrecognised aesthetic data in a layer. geom_line doesn't have a shape aesthetic, so the replicate column in the plot's main data frame is not filtered during the creation of the geom_line layer and is therefore a different length from the filtered data.
The solution is to create the plot like this:
v1 <- ggplot(d, aes(x, y, colour = treatment,
group = interaction(treatment, replicate)))
geom_point(aes(shape = replicate), size = 3)
geom_line()
So now you can do:
rm(replicate, treatment,x,y)
v1 gghighlight(replicate == "y")
and
v1 gghighlight(treatment == "a")