I have a fairly alrge dataset and I am running a for loop to remove one line per transect and calculate the frequency of the category. I am now trying to make it so that instead of one line per transect it removes a whole transect every iteration. Is it possible to do this?
Here is a sample dataset with the same columns I have
Transect<- c(1,1,1,1,1,2,2,2,2,2,3,3,3,3,3)
Category<- c("S","S","S","C","T","S","SP","T", "C", "T","S","SP","T","S","C")
dat<- data.frame(Transect,Category)
So the current code below removes one line per transect. How could I do it so that it randomly deletes a whole transect category (i.e. in the first iteration all of transect 3 is removed and on the second all of 1 is removed)
for (q in 1:2) {
for ( i in 0:5){
#if (i>0)
df<- dat2 %>%
group_by(Transect) %>%
sample_n(n() - i, replace = TRUE) %>%
ungroup()
c<-df %>%
group_by(Category) %>%
summarise(n = n(), replace=TRUE) %>%
mutate(freq = n / sum(n),
total=55-i)
if (i==0){
tot_1=c
} else {
tot_1=bind_rows(tot_1,c)
}
}
tot_1$rep = q
if (q==1){
dftot = tot_1
} else {
dftot=bind_rows(dftot, tot_1)
}
}
CodePudding user response:
It seems your goals is to iteratively assess increasingly small subsamples of your data to assess loss of representation of the whole. This code will try dropping a random 1 then 2 then 3... and report the distribution of categories. The last few lines normalize count to fraction of total for easy comparison between iterations.
Note I used set.seed() because it will return a different result each time due to random sampling.
To break down this answer a bit:
- It's important that
Categoryis afactorso thattable()won't drop anyCategoryvalues that have no count in a particular iteration. It would run to a point but then the rowbinding operation withinmap_dfr()would fail. - First I just enumerate the numbers of
Transectto leave out (should be 0:4 in this example) using0:length(unique(d$Transect)). I included 0 so that we can see what it looks like with the full dataset. - I used
set_names()so that it becomes a named vector. This allows us to use.idinsidemap_dfr()so that we get an extra column which stores the value of the leaveout. purrr::map_dfr()will iteratively apply a function over some list. In this case I piped in the list of leaveout values (which we just named) and the function we apply is given as an rlang-style lambda function which begins with~and operates on the argument.x.- Working from the inside of the
filteroperation, this function first randomly samples a number of values ofTransectto exclude given by.xand then removes data with said value ofTransect. Here we use%in%and negate the whole result with!at the beginning. - Then we just use
dplyr::pull()to take theCategorycolumn as avectorand runtable()on it to tabulate the occurrence of each value. - The rest just calculates the total count for each iteration and then divides the values by that to get a fraction.
library(tidyverse)
d <- tibble(
Transect = as.character(c(1,1,1,1,1,2,2,2,2,2,3,3,3,3,3)),
Category = factor(c("S","S","S","C","T","S","SP","T", "C", "T","S","SP","T","S","C"))
)
set.seed(1)
0:length(unique(d$Transect)) %>% set_names() %>%
map_dfr( ~ d %>%
filter(!Transect %in% sample(unique(d$Transect), size = .x)) %>%
pull(Category) %>%
table(),
.id = "leaveout_transects") %>%
rowwise() %>%
mutate(total_count = sum(c_across(-1)), .after = 1) %>%
mutate(across(-c(1:2), ~.x/total_count))
#> # A tibble: 4 × 6
#> # Rowwise:
#> leaveout_transects total_count C S SP T
#> <chr> <int> <table> <table> <table> <table>
#> 1 0 15 0.2 0.4 0.1333333 0.2666667
#> 2 1 10 0.2 0.3 0.2000000 0.3000000
#> 3 2 5 0.2 0.2 0.2000000 0.4000000
#> 4 3 0 NaN NaN NaN NaN
It would probably be more rigorous to simulate each leaveout condition multiple times and look at the distribution of performance you get at each value to assess what's likely to happen in the future with a given subsample.
Base r has the built in function replicate which is great for this purpose. Here I'm just using the code above with replicate and then reformatting the data a bit to graph it.
# use replicate to make many simulations
n_reps <- 20
replicate(
n_reps,
0:length(unique(d$Transect)) %>% set_names() %>%
map_dfr(
~ d %>%
filter(!Transect %in% sample(unique(d$Transect), size = .x)) %>%
pull(Category) %>%
table(),
.id = "leaveout_transects"
) %>%
rowwise() %>%
mutate(total_count = sum(c_across(-1)), .after = 1) %>%
mutate(across(-c(1:2), ~ .x / total_count)) %>%
select(3:6) %>%
t() %>%
cor() %>%
.[, 1]) %>%
as_tibble(.name_repair = "unique") %>%
mutate("leavout_transects" = factor(0:length(unique(d$Transect)))) %>%
pivot_longer(-leavout_transects, values_to = "correlation") %>%
select(-name) %>%
ggplot(aes(leavout_transects, correlation))
geom_boxplot()
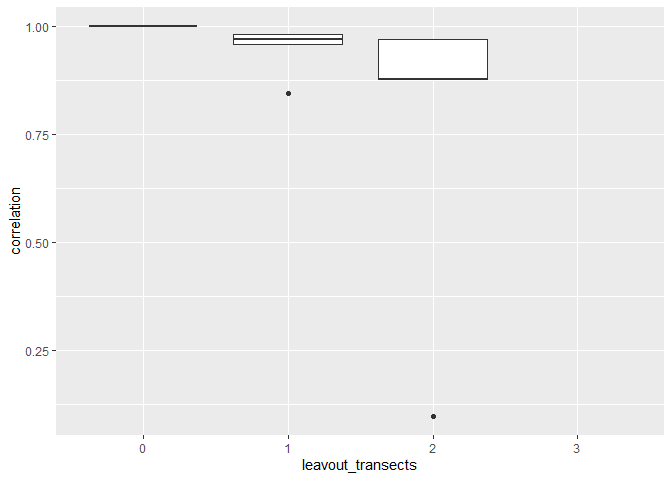
Created on 2022-09-22 by the reprex package (v2.0.1)
