I'm trying to change the borders of nodes in ggraph using geom_node_circle. I want to set the border colors based on a mapping between the names of the nodes and the colors that I supply for the names. I know that the parameter to change the border is colour.
Make graph for example:
df <- data_frame(group = c("animals","animals","pets","pets","pets","wild animals","wild animals"),
subgroup = c("pets","wild animals","rabbit","dog","cat","polar bear","panda bear"))
df <- as.data.frame(table(df))
df <- filter(df, Freq > 0)
vertices <- df %>%
dplyr::distinct(subgroup, Freq) %>%
dplyr::add_row(subgroup = "animals", Freq = 0)
graph <- graph_from_data_frame(df, vertices = vertices)
I know how to change all of the borders to a single new color (red):
ggraph(graph, layout = "circlepack", weight = Freq)
geom_node_circle(aes(fill =depth),colour = "red")
coord_fixed()
geom_node_label(aes(label = name), repel = TRUE, size = 5)
But, I don't know how to assign the border based on the mapping between name in graph and my colors.
Attempt A
Make map and apply to ggraph:
my_colors <- c("black","black","red","black","black","black","black","black")
names(my_colors) <- c("animals","pets","wild animals","rabbit","dog","cat","polar bear","panda bear")
ggraph(graph, layout = "circlepack", weight = Freq)
geom_node_circle(aes(fill =depth),colour = my_colors)
coord_fixed()
geom_node_label(aes(label = name), repel = TRUE, size = 5)
Returns
Error in `check_aesthetics()`:
! Aesthetics must be either length 1 or the same as the data (2888): colour
Run `rlang::last_error()` to see where the error occurred.
Attempt B
Add color column to original df and apply to ggraph:
df$my_colors <- cbind(df, ifelse(df$subgroup=="wild animals", "red", "black")
ggraph(graph, layout = "circlepack", weight = Freq)
geom_node_circle(aes(fill =depth),colour = df$my_colors)
coord_fixed()
geom_node_label(aes(label = name), repel = TRUE, size = 5)
Returns the same error as before.
When I look at the graph, I can see that my_colors isn't there:
create_layout(graph, 'circlepack', weight = Freq)
Non-leaf weights ignored
x y r circular leaf depth name Freq .ggraph.orig_index .ggraph.index
1 0.0000000 0.0000000 2.3440385 FALSE FALSE 0 animals 0 8 1
2 0.7244867 -0.8650770 1.2156595 FALSE FALSE 1 pets 1 4 2
3 -0.7805260 0.9319910 1.1283791 FALSE FALSE 1 wild animals 1 7 3
4 0.2584208 -0.4098895 0.5641896 FALSE TRUE 2 cat 1 1 4
5 1.3517237 -0.6890459 0.5641896 FALSE TRUE 2 dog 1 2 5
6 -0.5085709 0.4376730 0.5641896 FALSE TRUE 2 panda bear 1 3 6
7 -1.0524811 1.4263089 0.5641896 FALSE TRUE 2 polar bear 1 5 7
8 0.5633157 -1.4962957 0.5641896 FALSE TRUE 2 rabbit 1 6 8
Attempt C
Try adding my_color to vertices:
vertices$my_color <- ifelse(vertices$subgroup=="wild animals", "red", "black")
graph <- graph_from_data_frame(df, vertices = vertices)
# check if my_color is there
create_layout(graph, 'circlepack', weight = Freq)
I can see my_color is now in graph:
Non-leaf weights ignored
x y r circular leaf depth name Freq my_color .ggraph.orig_index .ggraph.index
1 0.00000000 0.0000000 2.3440387 FALSE FALSE 0 animals 0 black 8 1
2 0.42321874 1.0460045 1.2156596 FALSE FALSE 1 pets 1 black 4 2
3 -0.45595483 -1.1269132 1.1283791 FALSE FALSE 1 wild animals 1 red 7 3
4 1.05259805 1.2142151 0.5641896 FALSE TRUE 2 cat 1 black 1 4
5 0.25420381 0.4168406 0.5641896 FALSE TRUE 2 dog 1 black 2 5
6 -0.05740794 -0.7275760 0.5641896 FALSE TRUE 2 panda bear 1 black 3 6
7 -0.85450171 -1.5262505 0.5641896 FALSE TRUE 2 polar bear 1 black 5 7
8 -0.03714565 1.5069576 0.5641896 FALSE TRUE 2 rabbit 1 black 6 8
But if I run
ggraph(graph, layout = "circlepack", weight = Freq)
geom_node_circle(aes(fill =depth),colour = my_color)
coord_fixed()
geom_node_label(aes(label = name), repel = TRUE, size = 5)
I get
Error in layer(data = data, mapping = mapping, stat = StatNodeCircle, :
object 'my_color' not found
So, then I figured that I just don't know how to access attributes from graph but I figured that out and assigned it to the border color using colour = V(graph)$my_color. That still results in the same error indicating that the length of data is 2888. I assume that this error is being thrown because my list of colors is less than that.
Thoughts
I'm new to using ggraph, and I think that I'm lacking some fundamental knowledge to figure this out....
Solution
Modified from @DanAdams original answer:
df <- data_frame(group = c("animals","animals","pets","pets","pets","wild animals","wild animals"),
subgroup = c("pets","wild animals","rabbit","dog","cat","polar bear","panda bear"))
df <- as.data.frame(table(df))
df <- filter(df, Freq > 0)
vertices <- df %>%
dplyr::distinct(subgroup, Freq) %>%
dplyr::add_row(subgroup = "animals", Freq = 0)
vertices$my_color <- ifelse(vertices$subgroup=="wild animals", "red", "black")
graph <- graph_from_data_frame(df, vertices = vertices)
ggraph(graph, layout = "circlepack", weight = Freq)
geom_node_circle(aes(fill =depth,colour = my_color))
coord_fixed()
geom_node_label(aes(label = name), repel = TRUE, size = 5)
CodePudding user response:
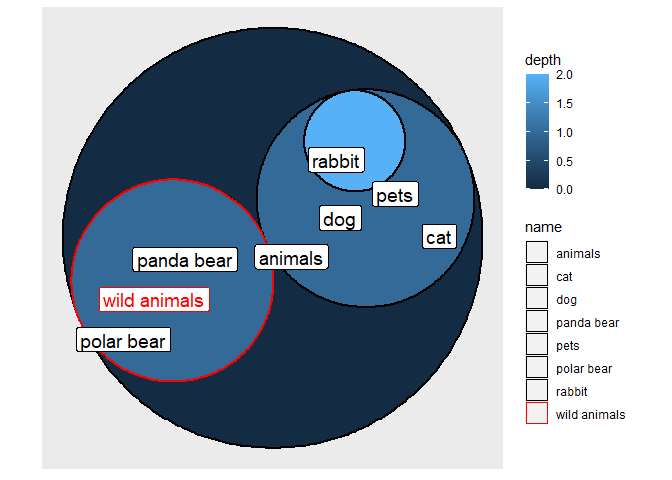
It seems you just need to add color = name inside of the aes().
I also colored the labels to match to make it easier to follow.
library(tidyverse)
library(igraph)
library(ggraph)
df <- data_frame(group = c("animals","animals","pets","pets","pets","wild animals","wild animals"),
subgroup = c("pets","wild animals","rabbit","dog","cat","polar bear","panda bear"))
my_colors <- c("black","black","red","black","black","black","black","black")
names(my_colors) <- c("animals","pets","wild animals","rabbit","dog","cat","polar bear","panda bear")
df <- as.data.frame(table(df))
df <- filter(df, Freq > 0)
vertices <- df %>%
dplyr::distinct(subgroup, Freq) %>%
dplyr::add_row(subgroup = "animals", Freq = 0)
graph <- graph_from_data_frame(df, vertices = vertices)
ggraph(graph, layout = "circlepack", weight = Freq)
geom_node_circle(aes(fill = depth, color = name), size = 1)
coord_fixed()
geom_node_label(aes(label = name, color = name), repel = TRUE, size = 5, show.legend = F)
scale_color_manual(values = my_colors)
Created on 2022-09-22 by the reprex package (v2.0.1)
