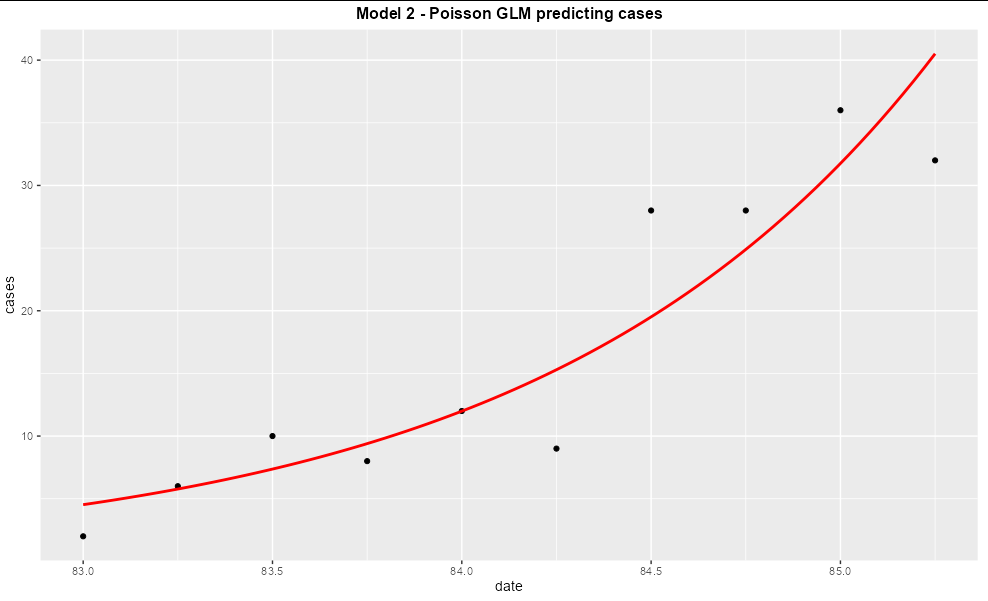
I have made a GLM and am trying to plot the model using ggplot using the following code. I think i need to add the type argument so that my model doesn't just produce a horizontal flat line. However i am struggling to fit it using the type argument as i run into this error message;
Error in model.frame.default(Terms, newdata, na.action = na.action, xlev =
object$xlevels) :
object is not a matrix
Here is my dataset and code used to get this if anyone knows the fix for this
my data (first 10 rows);
aids
cases quarter date
1 2 1 83.00
2 6 2 83.25
3 10 3 83.50
4 8 4 83.75
5 12 1 84.00
6 9 2 84.25
7 28 3 84.50
8 28 4 84.75
9 36 1 85.00
10 32 2 85.25
my code used to create the model and plot
model3 = glm(cases ~ date,
data = aids,
family = poisson(link='log'))
#plotting the model (“link”, “response”, “terms”)
plot_predictions <- function(model, type = 'response') {
#make predictions
preds <- predict(model, df)
#plot
df %>%
ggplot(aes(date, cases))
geom_point()
geom_line(aes(date, preds), col = 'red')
ggtitle("Model 2 - Poisson GLM predicting cases")
theme(plot.title = element_text(hjust = 0.5, size = 12, face = 'bold'))
}
plot_predictions(model3, aids)
dput output
dput(head(aids, 10))
structure(list(cases = c(2, 6, 10, 8, 12, 9, 28, 28, 36, 32),
quarter = structure(c(1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L,
2L), .Label = c("1", "2", "3", "4"), class = "factor"), date =
c(83,
83.25, 83.5, 83.75, 84, 84.25, 84.5, 84.75, 85, 85.25)),
row.names = c(NA,
10L), class = "data.frame")
CodePudding user response:
Here's one that puts the predictions on the right scale:
library(tidyverse)
aids <- structure(list(cases = c(2, 6, 10, 8, 12, 9, 28, 28, 36, 32),
quarter = structure(c(1L, 2L, 3L, 4L, 1L, 2L, 3L, 4L, 1L,
2L), .Label = c("1", "2", "3", "4"), class = "factor"), date =
c(83,
83.25, 83.5, 83.75, 84, 84.25, 84.5, 84.75, 85, 85.25)),
row.names = c(NA,
10L), class = "data.frame")
model3 = glm(cases ~ date,
data = aids,
family = poisson(link='log'))
plot_predictions <- function(model, df, type = 'response') {
require(tidyverse)
#make predictions
preds <- predict(model, df, type= type)
#plot
df %>%
ggplot(aes(date, cases))
geom_point()
geom_line(aes(date, preds), col = 'red')
ggtitle("Model 2 - Poisson GLM predicting cases")
theme(plot.title = element_text(hjust = 0.5, size = 12, face = 'bold'))
}
plot_predictions(model3, aids)
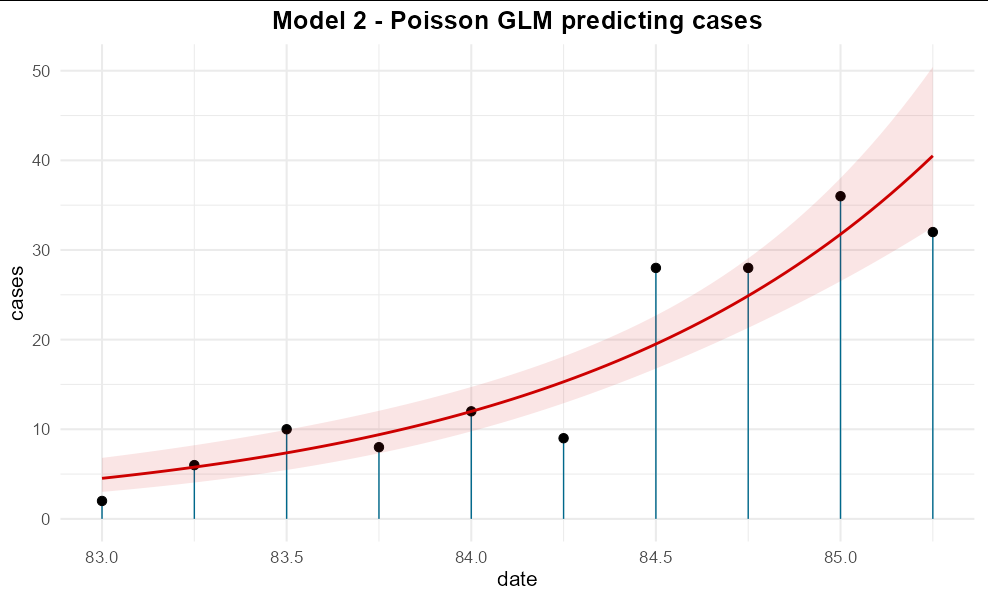
You can even get the standard error to show by omitting se = FALSE:
ggplot(aids, aes(date, cases))
geom_segment(aes(xend = date, yend = 0), color = "deepskyblue4")
geom_point(size = 3)
geom_smooth(col = 'red3', fill = "red3", method = glm, alpha = 0.1,
method.args = list(family = poisson(link = 'log')))
ggtitle("Model 2 - Poisson GLM predicting cases")
theme_minimal(base_size = 16)
theme(plot.title = element_text(hjust = 0.5, face = 'bold'))
CodePudding user response:
You can also do this using broom::augment to add the predictions of your model to the data and plot that:
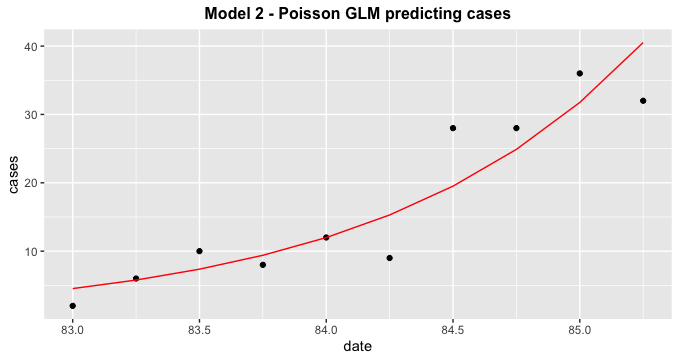
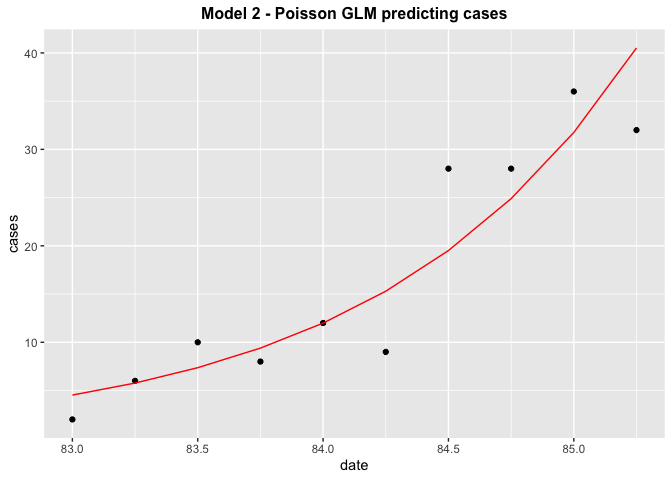
model3 %>%
broom::augment() %>%
ggplot(aes(date, cases))
geom_point()
geom_line(aes(date, exp(.fitted)), col = 'red')
ggtitle("Model 2 - Poisson GLM predicting cases")
theme(plot.title = element_text(hjust = 0.5, size = 12, face = 'bold'))