My algorithm is calculating an average over 1,000,000 separate processes, therefore uses Pool of python multiprocessing library (once I tried with map and once with imap)
When I a computer with 28, it did it with ~350 iterations/minute
When I moved to AWS EC2, which is with 192 CPU cores, it improved only by a factor of 2! ~700 iterations/minute.
I wonder if there is any good practice I am missing...?
I tried:
- monitor CPU usage - all of them seem to be used
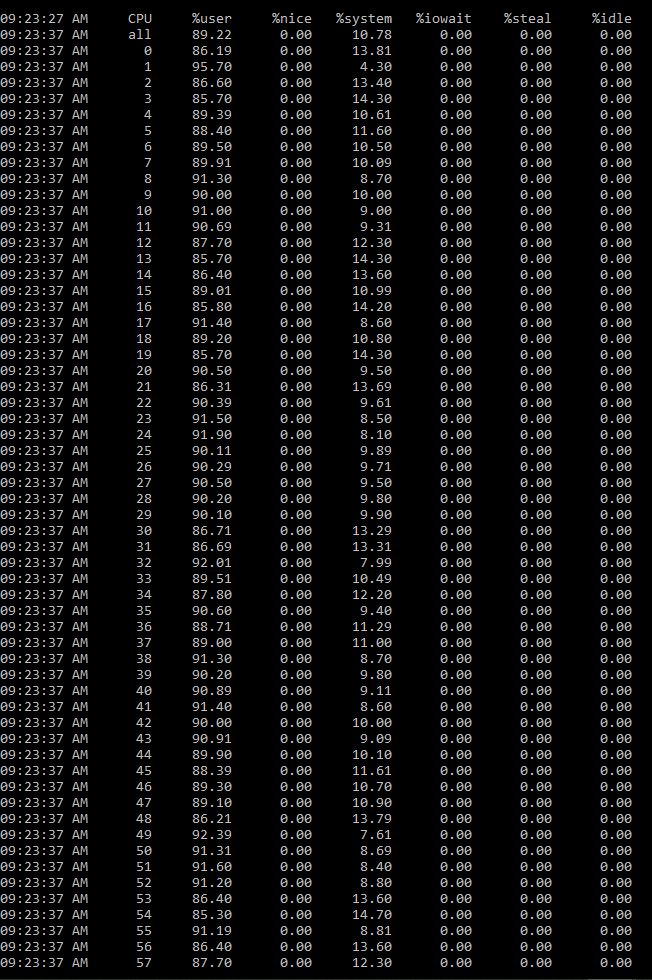
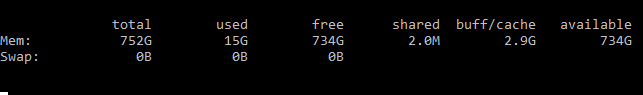
- Memory monitor - using only 15GB of 750GB RAM that the EC2 has
- using
mapinsteadimapdid not improve - seems that the sweet spot is giving
Poolparameter of 53 cores instead 192 (gives time of ~950 iter/min)
The code looks like:
import os
NUM_THREADS = "1"
os.environ["OMP_NUM_THREADS"] = NUM_THREADS
os.environ["OPENBLAS_NUM_THREADS"] = NUM_THREADS
os.environ["MKL_NUM_THREADS"] = NUM_THREADS
os.environ["VECLIB_MAXIMUM_THREADS"] = NUM_THREADS
os.environ["NUMEXPR_NUM_THREADS"] = NUM_THREADS
import numpy as np
import multiprocessing as mp
from multiprocessing import Pool
import time
def sample_in_pool(n_r_c_touple):
# calculations using matrices, multiplications, numpy - extensive use of cpu
return (xsyndrome, zsyndrome,
np.array(exact_choi_matrix),
np.array(correction_exact),
np.array(diamond_norms))
if __name__ == "__main__":
def run_test(rows,cols,nsamples, ncores):
starttime = time.time()
with Pool(processes=mp.cpu_count()) as pool:
result = pool.map(sample_in_pool, [(noise_pepso, rows, cols)] * nsamples)
xsyndrome = [x[0] for x in result]
zsyndrome = [x[1] for x in result]
exact_choi_matrix = [x[2] for x in result]
correction_exact = [x[3] for x in result]
diamond_norms = [x[4] for x in result]
pool.terminate()
pool.close()
samples_so_far = nsamples
"\nSamples/min (min): " str(round(samples_so_far / ((time.time() - starttime) / 60)))
def run_batch_of_d_N_composite(d, composite, N,nsamples):
for i in range(N):
diamond_mean, plot_str = run_test(rows=d, cols=d,
nsamples=nsamples, ncores=ncores,
noise_type="Composite",
rot_angle=0.1,
composite=composite, big_omega_error=False,
test_batch=test_batch_folder, computer_name=computer)
run_batch_of_d_N_composite(d=7, composite=False, N=1,nsamples=1500)
CodePudding user response:
First of all, multiprocessing is known to cause scalability issues due to the expensive inter-process communication (IPC). The only way to reduce this overhead is to reduce the amount of data transferred between processes (centralized slow operation) possibly by operating on shared memory. Multithreading can theoretically mitigate this problem but the Global Interpreter Lock (GIL) of the mainstream CPython interpreter prevents any speed-up unless it is released. This is the case for many Numpy operation but it will still not scale perfectly due to theoretical limitations (see later). If your computation only make Numpy operations, I strongly advise you not to use multiprocessing but the fork-join operations of Numba/Cython (they use multithreading but without the GIL issue so you do not pay the cost of creating process nor IPC).
Moreover, no computation can perfectly scale. The ones with a slight sequential portion of sequential work does not scale well on 192 cores. This is called the Amdahl's_law. Some resources likes the RAM is shared between cores and can be saturated (see this recent post for example).
Additionally, computing machine are not a set of independent cores, they are far more complex. Indeed, modern computing machines with a lot of cores are NUMA systems. On such machines, cores are grouped into NUMA nodes that can access efficiently to their own memory but less efficiently to the one of others. NUMA nodes haves their own memory that can be easily saturated if all the memory is allocated on the same node. Thus, you need to balance data on several nodes so to avoid this problem. This is done by controlling the NUMA allocation policy. The default policy is generally to perform a local allocation during the first-touch on a memory page so you need to perform local accesses. Multiprocessing tends to have a good behaviour regarding the allocation policy assuming processes do not moves between cores. Thus, you need to bind processes to cores so to improve data locality. Further, more physical cores contains multiple hardware threads sharing the same resources (so it is generally not much faster to run 2 processes on the same core if they are doing optimized numerically-intensive operations). This is called SMT and it is useful to speed up latency bound computations (eg. IO-bound codes, data transpositions, unoptimized scalar codes). This is a very complex topic. For more information please read this page and possibly high-performance computing books.
In fact, EC2 machines have 2 sockets of AMD EPYC 7R13 with 48 core each and 2 threads per cores. This is a good example of complex NUMA architecture with SMT. Thus, in practice, you should expect only a speed up smaller than 96 (assuming the IPC does not help much which is unlikely for Numpy codes). Because Numpy perform a lot of temporary array copies, the code can certainly be memory bound and saturate the RAM (see this post or this one for example). Again, Numba/Cython can help to reduce the number of temporary array created and make the whole computation less memory-bound (and thus eventually scale better).
CodePudding user response:
There could be many possible reasons why your code might not be performing as well as you'd want it to, but in this answer I will focus on one such area: data serialization in multiprocessing.
Whenever you create a pool, all the arguments need to be serialized so that they can be used with another process (also called pickling). Multiprocessing uses pickle to do so. This serialization happens for every task that the pool creates, rather than per process. For example, consider the code below, where we subclass the builtin str class and override the dunder methods __getstate__ and __setstate__. These methods are used to tell python (and pickle) how to serialize/deserialize your object. We leave a helpful print statement to know what is happening when we create our pool:
class NewString(str):
def __getstate__(self):
print('being serialized')
return self.__dict__
def __setstate__(self, state):
print('being deserialized?')
self.__dict__ = state
def worker(string):
print(string)
if __name__ == "__main__":
p = Pool(2)
string = NewString('inside worker')
args = [string for _ in range(5)]
time.sleep(5)
p.map(worker, args)
Output
being serialized
being serialized
being serialized
being serialized
being serialized
being deserialized?
inside worker
being deserialized?
inside worker
being deserialized?
inside worker
being deserialized?
inside worker
being deserialized?
inside worker
As you can see, there were a total of 5 pickling, and 5 unpickling calls made, even though the number of processes the pool was started with was 2. Again, this is because the Pool class pickles these arguments per task, rather than per worker.
This obviously adds extra-overhead, which scales with the complexity of the objects that need to pickled (keep in mind the result from the worker is pickled as well). Therefore, if you are passing "big matrices, with a size in million of numbers", and creating a million tasks, these extra pickling/unpickling calls will take longer as well (more complex objects), and impact your performance.
So what's the solution?
Ideally, we want to restrict the number of pickling calls to at most the number of worker we create (below that would be impossible). To do this, we use multiprocessing.Process instead of Pool (see this answer for detailed demonstration of only pickling once per worker using process). The gist of the approach here is that instead of creating a few workers and passing them a lot of tasks using a pool, we create a few intermediatory workers using Process, and pass them the list of tasks which they can execute using the original workers in their own process, without needing to pickle/unpickle. This is made much clearer in the code below. But before that, coming back to your use-case, since the tasks are more cpu-bound, lets assume the optimal number of processes we can create is 8 (this will be different for your machine). Therefore, if we can limit the number of serialization calls to 8, then exactly how much of a difference can we expect? Let's check using the code below:
import multiprocessing
from multiprocessing import Pool, Process
import random
import time
def main_worker(my_list):
new_list = random.sample(range(10000, 99999), 1000)
return new_list
def indirect_worker(count, arg):
t = time.time()
results = []
for i in range(count):
results.append(main_worker(arg[i]))
print(f"Process completed in : {time.time() - t}")
if __name__ == "__main__":
num_proc = 8
length = 80000
num_args = length//num_proc
args = [random.sample(range(10000, 99999), length)] * length
p = Pool(num_proc)
t = time.time()
p.map(main_worker, args)
print(f'Pool completed in : {time.time() - t}')
processes = []
for i in range(8):
# In this, i * num_args is the starting index and i * num_args num_args is the ending index of arguments for
# to provide to this process
processes.append(Process(target=indirect_worker, args=(num_args, args[i * num_args:i * num_args num_args])))
processes[-1].start()
for process in processes:
process.join()
In this code, we first use a Pool of 8 processes to complete 80,000 tasks. The actual task is executed by main_worker, which just returns an arbitrary list of a 1000 numbers. An arbitrary list my_list is also passed to the main_worker as the argument. The only thing relevant about this list is that it's a fairly large list of 80,000 numbers, which was my attempt to make the data needed to be pickled larger. After the pool finishes executing it's tasks, it now uses the same argument list args used for the pool (a list of 80,000 lists of 80,000 numbers) to start 8 indirect_workers using multiprocessing.Process. Each indirect_worker recieves of portion of args which it uses to start the main_worker. Therefore, by using the "process" setup we are only creating 8 workers, and hence only pickling/unpickling 8 times.
Here is the result of our test:
Pool completed in : 16.491206169128418
Process completed in : 10.309278726577759
Process completed in : 10.292876482009888
Process completed in : 10.574358701705933
Process completed in : 10.27439832687378
Process completed in : 10.180659055709839
Process completed in : 10.588935613632202
Process completed in : 10.58601999282837
Process completed in : 10.707130193710327
Now keep in mind that we had 8 processes running concurrently, therefore there are 8 prints for them. Assuming the processes took 11.0s (a little time should be added since starting the process itself wasn't accounted for) to start and finish, that leaves us with a straight 33% improvement in speed. Keep in mind however, that the actual improvement will be lesser if you're not using shared memory, since in the indirect_worker we were not pickling the results to send to the main process. With that said, this improvement was done with fairly simple objects as arguments which needed to be pickled/unpickled, and I assume it will scale as the complexity of the arguments grow. In fact, if it's feasible for your structure, you can actually use multiprocessing.shared_memory with numpy in indirect_worker to store the return values and get even better speed benefits over pool's built in serialization of return values since you won't even have to pickle/unpickle them at all!
Note: I noticed this line in your code in run_test:
result = pool.map(sample_in_pool, [(noise_pepso, rows, cols)] * nsamples)
Even though your RAM usage isn't very high, it's probably better to use generators as the iterable rather than storing copies of the same object in memory
