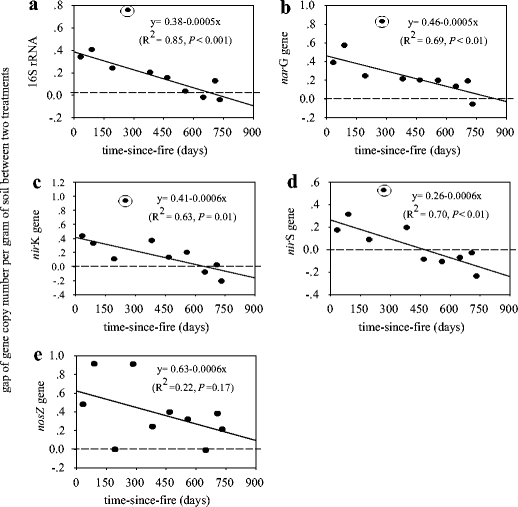
I want to perform linear regression analysis to determine the effect of time-dependent treatment on gene expression and survival for each gene.
In df4 dataframe, the time column represents time, the last column survival_info represents survival info, and the remaining columns represent gene expression.
For each row of df5 dataframe, I want to individually fit, predict and plot linear regression using sklearn.
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
df5 = df4.groupby(["time"], sort=False).mean()
df5 = df5.reset_index()
df5["time"] = df5["time"].str[:-1]
df5 = df5.set_index("time").T
# split the dataset into train and test sets
X = df5.iloc[:,1:]
y = df5.iloc[:,0]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# Linear regression analysis
reg = LinearRegression().fit(X_train, y_train)
y_pred = reg.predict(X_test)
# Plot
for i in X_test.index:
fig = px.scatter(X_test.loc[i], y_pred.flatten())
fig.update_layout(title="Regression analysis of time-dependent treatment on " i " expression", xaxis_title="Time", yaxis_title="Gene Expression", legend_title="Survival info")
fig.show()
Traceback:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-422-04ea16d131bd> in <module>()
1 # Plot
2 for i in X_test.index:
----> 3 fig = px.scatter(X_test.loc[i], y_pred.flatten())
4 fig.update_layout(title="Regression analysis of time-dependent treatment on " i " expression", xaxis_title="Time", yaxis_title="Gene Expression", legend_title="Survival info")
5 fig.show()
3 frames
/usr/local/lib/python3.7/dist-packages/plotly/express/_chart_types.py in scatter(data_frame, x, y, color, symbol, size, hover_name, hover_data, custom_data, text, facet_row, facet_col, facet_col_wrap, facet_row_spacing, facet_col_spacing, error_x, error_x_minus, error_y, error_y_minus, animation_frame, animation_group, category_orders, labels, orientation, color_discrete_sequence, color_discrete_map, color_continuous_scale, range_color, color_continuous_midpoint, symbol_sequence, symbol_map, opacity, size_max, marginal_x, marginal_y, trendline, trendline_options, trendline_color_override, trendline_scope, log_x, log_y, range_x, range_y, render_mode, title, template, width, height)
64 mark in 2D space.
65 """
---> 66 return make_figure(args=locals(), constructor=go.Scatter)
67
68
/usr/local/lib/python3.7/dist-packages/plotly/express/_core.py in make_figure(args, constructor, trace_patch, layout_patch)
1943 apply_default_cascade(args)
1944
-> 1945 args = build_dataframe(args, constructor)
1946 if constructor in [go.Treemap, go.Sunburst, go.Icicle] and args["path"] is not None:
1947 args = process_dataframe_hierarchy(args)
/usr/local/lib/python3.7/dist-packages/plotly/express/_core.py in build_dataframe(args, constructor)
1404
1405 df_output, wide_id_vars = process_args_into_dataframe(
-> 1406 args, wide_mode, var_name, value_name
1407 )
1408
/usr/local/lib/python3.7/dist-packages/plotly/express/_core.py in process_args_into_dataframe(args, wide_mode, var_name, value_name)
1247 "The length of argument `%s` is %d, whereas the "
1248 "length of previously-processed arguments %s is %d"
-> 1249 % (field, len(argument), str(list(df_output.columns)), length)
1250 )
1251 df_output[str(col_name)] = to_unindexed_series(argument)
ValueError: All arguments should have the same length. The length of argument `y` is 5, whereas the length of previously-processed arguments ['x'] is 18
Data:
df4.head().to_dict()
{'DNAJA1': {'0h_T1_TimeC1_PIDC1_Survivor': 0.30466896848826,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.378391959798995,
'0h_T1_TimeC2_PIDC2_Survivor': 0.2466734902763562,
'0h_T1_TimeC4_PIDC3_Survivor': 0.20020120724346072,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.21559633027522937},
'DNAJA1P5': {'0h_T1_TimeC1_PIDC1_Survivor': -0.1971955281717348,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.19145728643216084,
'0h_T1_TimeC2_PIDC2_Survivor': -0.2231320368474923,
'0h_T1_TimeC4_PIDC3_Survivor': -0.22032193158953728,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.21457696228338433},
'DNAJA2': {'0h_T1_TimeC1_PIDC1_Survivor': 0.25586022467105374,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.22261306532663308,
'0h_T1_TimeC2_PIDC2_Survivor': 0.25588536335721596,
'0h_T1_TimeC4_PIDC3_Survivor': 0.21730382293762568,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.17380224260958205},
'DNAJA3': {'0h_T1_TimeC1_PIDC1_Survivor': 0.12484533092566467,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.18040201005025125,
'0h_T1_TimeC2_PIDC2_Survivor': 0.12384851586489252,
'0h_T1_TimeC4_PIDC3_Survivor': 0.13380281690140847,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.16666666666666663},
'DNAJA4': {'0h_T1_TimeC1_PIDC1_Survivor': 0.25447309843039345,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.45477386934673364,
'0h_T1_TimeC2_PIDC2_Survivor': 0.20573183213920163,
'0h_T1_TimeC4_PIDC3_Survivor': 0.25955734406438624,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2706422018348623},
'DNAJB1': {'0h_T1_TimeC1_PIDC1_Survivor': 0.36540720966934437,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.5793969849246231,
'0h_T1_TimeC2_PIDC2_Survivor': 0.3316274309109519,
'0h_T1_TimeC4_PIDC3_Survivor': 0.30281690140845063,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.41743119266055034},
'DNAJB11': {'0h_T1_TimeC1_PIDC1_Survivor': 0.33305451443172285,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.31005025125628144,
'0h_T1_TimeC2_PIDC2_Survivor': 0.21596724667349024,
'0h_T1_TimeC4_PIDC3_Survivor': 0.23943661971830987,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.23802242609582047},
'DNAJB12': {'0h_T1_TimeC1_PIDC1_Survivor': 0.1304748089195234,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.08492462311557787,
'0h_T1_TimeC2_PIDC2_Survivor': 0.12077789150460601,
'0h_T1_TimeC4_PIDC3_Survivor': 0.09054325955734402,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.10448521916411818},
'DNAJB13': {'0h_T1_TimeC1_PIDC1_Survivor': -0.06551009305018526,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.04773869346733674,
'0h_T1_TimeC2_PIDC2_Survivor': -0.08904810644831117,
'0h_T1_TimeC4_PIDC3_Survivor': -0.10462776659959759,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.06472986748216113},
'DNAJB14': {'0h_T1_TimeC1_PIDC1_Survivor': 0.31506844602697626,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.27487437185929653,
'0h_T1_TimeC2_PIDC2_Survivor': 0.2947799385875129,
'0h_T1_TimeC4_PIDC3_Survivor': 0.289738430583501,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.27268093781855235},
'DNAJB2': {'0h_T1_TimeC1_PIDC1_Survivor': 0.20049372379622737,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.18643216080402006,
'0h_T1_TimeC2_PIDC2_Survivor': 0.12998976458546577,
'0h_T1_TimeC4_PIDC3_Survivor': 0.11569416498993959,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.15239551478083588},
'DNAJB3': {'0h_T1_TimeC1_PIDC1_Survivor': -0.06546246185535144,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.09396984924623121,
'0h_T1_TimeC2_PIDC2_Survivor': -0.10133060388945755,
'0h_T1_TimeC4_PIDC3_Survivor': -0.12273641851106647,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.09531090723751279},
'DNAJB4': {'0h_T1_TimeC1_PIDC1_Survivor': 0.06816950142840666,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.26381909547738686,
'0h_T1_TimeC2_PIDC2_Survivor': -0.0071647901740019854,
'0h_T1_TimeC4_PIDC3_Survivor': -0.014084507042253579,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.02599388379204892},
'DNAJB5': {'0h_T1_TimeC1_PIDC1_Survivor': 0.08117546379552881,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.12311557788944721,
'0h_T1_TimeC2_PIDC2_Survivor': 0.08802456499488233,
'0h_T1_TimeC4_PIDC3_Survivor': 0.08752515090543261,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.12181447502548413},
'DNAJB6 /// TMEM135': {'0h_T1_TimeC1_PIDC1_Survivor': 0.27126097766732055,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.31909547738693467,
'0h_T1_TimeC2_PIDC2_Survivor': 0.3070624360286592,
'0h_T1_TimeC4_PIDC3_Survivor': 0.3078470824949698,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.20948012232415897},
'DNAJB7': {'0h_T1_TimeC1_PIDC1_Survivor': -0.13556605441176003,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.13015075376884422,
'0h_T1_TimeC2_PIDC2_Survivor': -0.17297850562947795,
'0h_T1_TimeC4_PIDC3_Survivor': -0.19215291750503022,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.14424057084607542},
'DNAJB8': {'0h_T1_TimeC1_PIDC1_Survivor': -0.05724766845301285,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.042713567839195964,
'0h_T1_TimeC2_PIDC2_Survivor': -0.06243602865916064,
'0h_T1_TimeC4_PIDC3_Survivor': -0.07042253521126762,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.063710499490316},
'DNAJB9': {'0h_T1_TimeC1_PIDC1_Survivor': 0.1658875438074684,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.17135678391959802,
'0h_T1_TimeC2_PIDC2_Survivor': 0.07062436028659165,
'0h_T1_TimeC4_PIDC3_Survivor': 0.03521126760563377,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.07186544342507646},
'DNAJC1': {'0h_T1_TimeC1_PIDC1_Survivor': 0.2780145518592573,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.23567839195979898,
'0h_T1_TimeC2_PIDC2_Survivor': 0.24257932446264086,
'0h_T1_TimeC4_PIDC3_Survivor': 0.23943661971830987,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.24006116207951073},
'DNAJC10': {'0h_T1_TimeC1_PIDC1_Survivor': 0.25124370342066066,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.28190954773869353,
'0h_T1_TimeC2_PIDC2_Survivor': 0.23029682702149437,
'0h_T1_TimeC4_PIDC3_Survivor': 0.25754527162977864,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.27166156982670736},
'DNAJC11': {'0h_T1_TimeC1_PIDC1_Survivor': 0.0779460687857962,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.14221105527638192,
'0h_T1_TimeC2_PIDC2_Survivor': 0.06448311156601842,
'0h_T1_TimeC4_PIDC3_Survivor': 0.11569416498993959,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.1564729867482161},
'DNAJC12': {'0h_T1_TimeC1_PIDC1_Survivor': -0.09891279151614327,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.11507537688442217,
'0h_T1_TimeC2_PIDC2_Survivor': -0.1361310133060389,
'0h_T1_TimeC4_PIDC3_Survivor': -0.14185110663983905,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.13812436289500513},
'DNAJC13': {'0h_T1_TimeC1_PIDC1_Survivor': 0.3859045005128292,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.35829145728643225,
'0h_T1_TimeC2_PIDC2_Survivor': 0.42988741044012296,
'0h_T1_TimeC4_PIDC3_Survivor': 0.4295774647887325,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.35524974515800195},
'DNAJC14': {'0h_T1_TimeC1_PIDC1_Survivor': 0.25440959017061504,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.23768844221105523,
'0h_T1_TimeC2_PIDC2_Survivor': 0.2128966223132037,
'0h_T1_TimeC4_PIDC3_Survivor': 0.1971830985915493,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2543323139653414},
'DNAJC15': {'0h_T1_TimeC1_PIDC1_Survivor': 0.3008182510036951,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.2597989949748744,
'0h_T1_TimeC2_PIDC2_Survivor': 0.24257932446264086,
'0h_T1_TimeC4_PIDC3_Survivor': 0.22837022132796786,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2635066258919469},
'DNAJC16': {'0h_T1_TimeC1_PIDC1_Survivor': 0.23949414612615494,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.25778894472361813,
'0h_T1_TimeC2_PIDC2_Survivor': 0.2589559877175025,
'0h_T1_TimeC4_PIDC3_Survivor': 0.24849094567404426,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2737003058103976},
'DNAJC17': {'0h_T1_TimeC1_PIDC1_Survivor': 0.05949056849418747,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.08793969849246232,
'0h_T1_TimeC2_PIDC2_Survivor': 0.060388945752302955,
'0h_T1_TimeC4_PIDC3_Survivor': 0.07947686116700202,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.0922528032619775},
'DNAJC18': {'0h_T1_TimeC1_PIDC1_Survivor': 0.06714595997497777,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.0809045226130653,
'0h_T1_TimeC2_PIDC2_Survivor': 0.03787103377686798,
'0h_T1_TimeC4_PIDC3_Survivor': 0.026156941649899377,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.09836901121304789},
'DNAJC19': {'0h_T1_TimeC1_PIDC1_Survivor': -0.056208249934639354,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.028643216080402028,
'0h_T1_TimeC2_PIDC2_Survivor': -0.08597748208802455,
'0h_T1_TimeC4_PIDC3_Survivor': -0.07444668008048293,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.05861365953109074},
'DNAJC2': {'0h_T1_TimeC1_PIDC1_Survivor': 0.13301249313316946,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.1522613065326633,
'0h_T1_TimeC2_PIDC2_Survivor': 0.09825997952917093,
'0h_T1_TimeC4_PIDC3_Survivor': 0.1106639839034205,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.14424057084607542},
'DNAJC21': {'0h_T1_TimeC1_PIDC1_Survivor': 0.19769142183350463,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.20954773869346735,
'0h_T1_TimeC2_PIDC2_Survivor': 0.1463664278403276,
'0h_T1_TimeC4_PIDC3_Survivor': 0.1891348088531187,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.19520897043832813},
'DNAJC22': {'0h_T1_TimeC1_PIDC1_Survivor': -0.07788256052601769,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.05879396984924623,
'0h_T1_TimeC2_PIDC2_Survivor': -0.11463664278403277,
'0h_T1_TimeC4_PIDC3_Survivor': -0.12173038229376258,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.0922528032619776},
'DNAJC24': {'0h_T1_TimeC1_PIDC1_Survivor': -0.04031160327660278,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.028643216080402028,
'0h_T1_TimeC2_PIDC2_Survivor': -0.029682702149437058,
'0h_T1_TimeC4_PIDC3_Survivor': 0.0010060362173038016,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.027013251783894042},
'DNAJC25 /// DNAJC25-GNG10 /// GNG10': {'0h_T1_TimeC1_PIDC1_Survivor': 0.2051155374016019,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.19145728643216084,
'0h_T1_TimeC2_PIDC2_Survivor': 0.1924257932446265,
'0h_T1_TimeC4_PIDC3_Survivor': 0.16700201207243454,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.17787971457696228},
'DNAJC27': {'0h_T1_TimeC1_PIDC1_Survivor': 0.06399595028996814,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.04974874371859298,
'0h_T1_TimeC2_PIDC2_Survivor': 0.0051177072671443015,
'0h_T1_TimeC4_PIDC3_Survivor': 0.009054325955734393,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.08307849133537201},
'DNAJC28': {'0h_T1_TimeC1_PIDC1_Survivor': -0.10060158199075109,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.06482412060301512,
'0h_T1_TimeC2_PIDC2_Survivor': -0.12691914022517914,
'0h_T1_TimeC4_PIDC3_Survivor': -0.13179074446680086,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.09021406727828753},
'DNAJC3': {'0h_T1_TimeC1_PIDC1_Survivor': 0.2834111662339285,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.2648241206030152,
'0h_T1_TimeC2_PIDC2_Survivor': 0.3643807574206756,
'0h_T1_TimeC4_PIDC3_Survivor': 0.2977867203219316,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.19317023445463807},
'DNAJC30': {'0h_T1_TimeC1_PIDC1_Survivor': 0.11612405915159318,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.12713567839195977,
'0h_T1_TimeC2_PIDC2_Survivor': 0.11873080859774823,
'0h_T1_TimeC4_PIDC3_Survivor': 0.09356136820925551,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.13608562691131498},
'DNAJC4 /// NUDT22': {'0h_T1_TimeC1_PIDC1_Survivor': 0.24217472392430245,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.21959798994974872,
'0h_T1_TimeC2_PIDC2_Survivor': 0.2906857727737973,
'0h_T1_TimeC4_PIDC3_Survivor': 0.22032193158953728,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2359836901121304},
'DNAJC5': {'0h_T1_TimeC1_PIDC1_Survivor': 0.3032077492778582,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.242713567839196,
'0h_T1_TimeC2_PIDC2_Survivor': 0.3285568065506654,
'0h_T1_TimeC4_PIDC3_Survivor': 0.295774647887324,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.25535168195718644},
'DNAJC5B': {'0h_T1_TimeC1_PIDC1_Survivor': -0.16639402217920127,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.1482412060301508,
'0h_T1_TimeC2_PIDC2_Survivor': -0.1668372569089048,
'0h_T1_TimeC4_PIDC3_Survivor': -0.17907444668008052,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.15545361875637106},
'DNAJC5G': {'0h_T1_TimeC1_PIDC1_Survivor': -0.11752177010221651,
'0h_T1_TimeC1_PIDC4_Non-Survivor': -0.09095477386934676,
'0h_T1_TimeC2_PIDC2_Survivor': -0.11873080859774823,
'0h_T1_TimeC4_PIDC3_Survivor': -0.15895372233400404,
'4h_T2_TimeC1_PIDC4_Non-Survivor': -0.11671763506625896},
'DNAJC6': {'0h_T1_TimeC1_PIDC1_Survivor': 0.044744479809136525,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.03165829145728639,
'0h_T1_TimeC2_PIDC2_Survivor': -0.008188331627430918,
'0h_T1_TimeC4_PIDC3_Survivor': -0.014084507042253579,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.008664627930682973},
'DNAJC7': {'0h_T1_TimeC1_PIDC1_Survivor': 0.24253301635655233,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.2175879396984925,
'0h_T1_TimeC2_PIDC2_Survivor': 0.2128966223132037,
'0h_T1_TimeC4_PIDC3_Survivor': 0.19919517102615691,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.1819571865443425},
'DNAJC8': {'0h_T1_TimeC1_PIDC1_Survivor': 0.14284727639535588,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.14623115577889448,
'0h_T1_TimeC2_PIDC2_Survivor': 0.11668372569089054,
'0h_T1_TimeC4_PIDC3_Survivor': 0.1277665995975855,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.11875637104994903},
'DNAJC9': {'0h_T1_TimeC1_PIDC1_Survivor': 0.10419456178771519,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.15527638190954773,
'0h_T1_TimeC2_PIDC2_Survivor': 0.09518935516888441,
'0h_T1_TimeC4_PIDC3_Survivor': 0.09356136820925551,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.11162079510703361},
'LOC646358': {'0h_T1_TimeC1_PIDC1_Survivor': 0.24757133829897363,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.21959798994974872,
'0h_T1_TimeC2_PIDC2_Survivor': 0.25588536335721596,
'0h_T1_TimeC4_PIDC3_Survivor': 0.22434607645875249,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.21865443425076447},
'ST13': {'0h_T1_TimeC1_PIDC1_Survivor': 0.4051681434101184,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.45577889447236175,
'0h_T1_TimeC2_PIDC2_Survivor': 0.34288638689866935,
'0h_T1_TimeC4_PIDC3_Survivor': 0.3581488933601609,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.4001019367991844},
'ST13P4': {'0h_T1_TimeC1_PIDC1_Survivor': 0.22716930984515626,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.2507537688442211,
'0h_T1_TimeC2_PIDC2_Survivor': 0.17604912998976466,
'0h_T1_TimeC4_PIDC3_Survivor': 0.1961770623742455,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.21457696228338433},
'ST13P5': {'0h_T1_TimeC1_PIDC1_Survivor': 0.26162201153945086,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.28190954773869353,
'0h_T1_TimeC2_PIDC2_Survivor': 0.19856704196519956,
'0h_T1_TimeC4_PIDC3_Survivor': 0.21327967806841047,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2420998980632008},
'VEGFA': {'0h_T1_TimeC1_PIDC1_Survivor': 0.3142407217078641,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.2668341708542714,
'0h_T1_TimeC2_PIDC2_Survivor': 0.11668372569089054,
'0h_T1_TimeC4_PIDC3_Survivor': 0.09154929577464782,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.2319062181447503},
'VEGFB': {'0h_T1_TimeC1_PIDC1_Survivor': 0.124370606683821,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.19547738693467331,
'0h_T1_TimeC2_PIDC2_Survivor': 0.14943705220061412,
'0h_T1_TimeC4_PIDC3_Survivor': 0.1006036217303823,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.19011213047910297},
'VEGFC': {'0h_T1_TimeC1_PIDC1_Survivor': 0.008064755138612082,
'0h_T1_TimeC1_PIDC4_Non-Survivor': 0.028643216080402028,
'0h_T1_TimeC2_PIDC2_Survivor': 0.012282497441146378,
'0h_T1_TimeC4_PIDC3_Survivor': -0.011066398390342085,
'4h_T2_TimeC1_PIDC4_Non-Survivor': 0.015800203873598304},
'survival_info': {'0h_T1_TimeC1_PIDC1_Survivor': 'Survivor',
'0h_T1_TimeC1_PIDC4_Non-Survivor': 'Non-Survivor',
'0h_T1_TimeC2_PIDC2_Survivor': 'Survivor',
'0h_T1_TimeC4_PIDC3_Survivor': 'Survivor',
'4h_T2_TimeC1_PIDC4_Non-Survivor': 'Non-Survivor'},
'time': {'0h_T1_TimeC1_PIDC1_Survivor': '0h',
'0h_T1_TimeC1_PIDC4_Non-Survivor': '0h',
'0h_T1_TimeC2_PIDC2_Survivor': '0h',
'0h_T1_TimeC4_PIDC3_Survivor': '0h',
'4h_T2_TimeC1_PIDC4_Non-Survivor': '4h'}}
I want to achieve something similar to this:
CodePudding user response:
As per your code:
X = pd.DataFrame(df5.loc[i]).iloc[1:,]
y = pd.DataFrame(df5.loc[i]).iloc[0,]
So X is n dimensional where as y is a scalar.
However in linear regression, given a set of features (called independent variables) per sample and a corresponding prediction (called dependent variable), you fit a model such that the models prediction of the dependent variable and the actual value is minimised for all the samples.
So you will need a set of samples, each sample represented by a set of features and a corresponding prediction for that sample.
Or just simply X.shape[0] == y.shape[0]
and X.shape == (n,m) and y.shape == (n,)
where n: No:of Samples, m: No:of features per sample
CodePudding user response:
Your for loop is iterating over the wrong values. The expression
for i in X_test.index:
...
iterates over the rows of X_test. However, it's the columns of X_test that should have the same length as y_pred.
This may get you closer to what you want:
for col_name in X_test.columns:
fig = px.scatter(X_test[col_name], y_pred.flatten())
fig.update_layout(title=("Regression analysis of time-dependent treatment on " i
" expression"),
xaxis_title="Time",
yaxis_title="Gene Expression",
legend_title="Survival info")
fig.show()
Depending on what you are using to plot your data, this may just create a bunch of overlapping plots, though, rather than a set of subplots.
Judging from this question and your previous one, it looks like you don't have a very good grasp of Python or the various packages that you are using. I strongly recommend that find some good textbooks and/or tutorials to help you get a sense of how they work. Also, remember that while Stack Overflow may help you if you're stuck, it isn't a code-writing service.