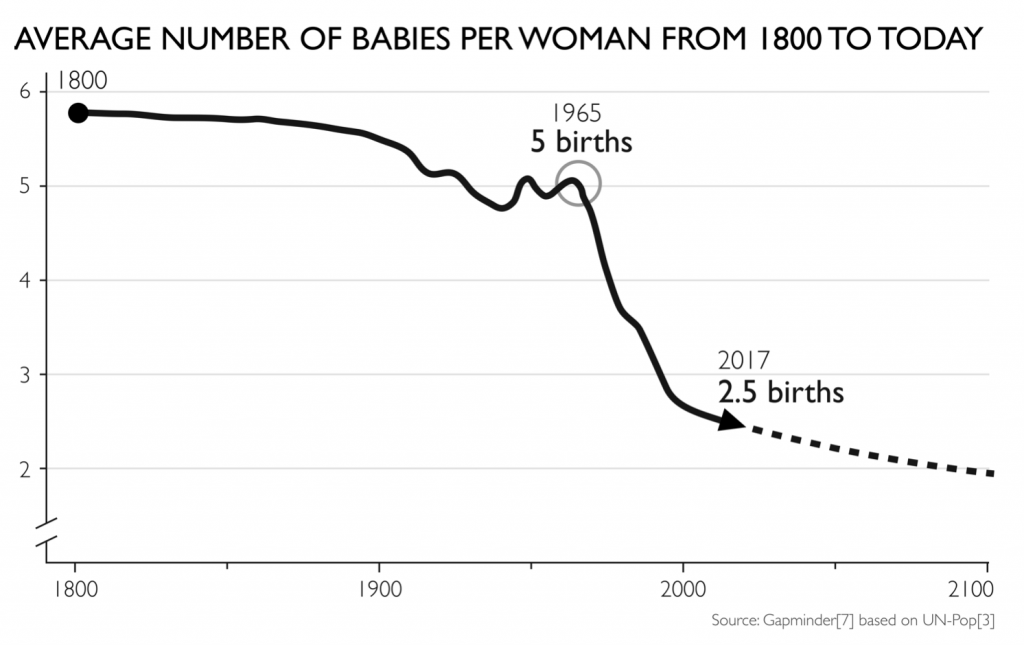
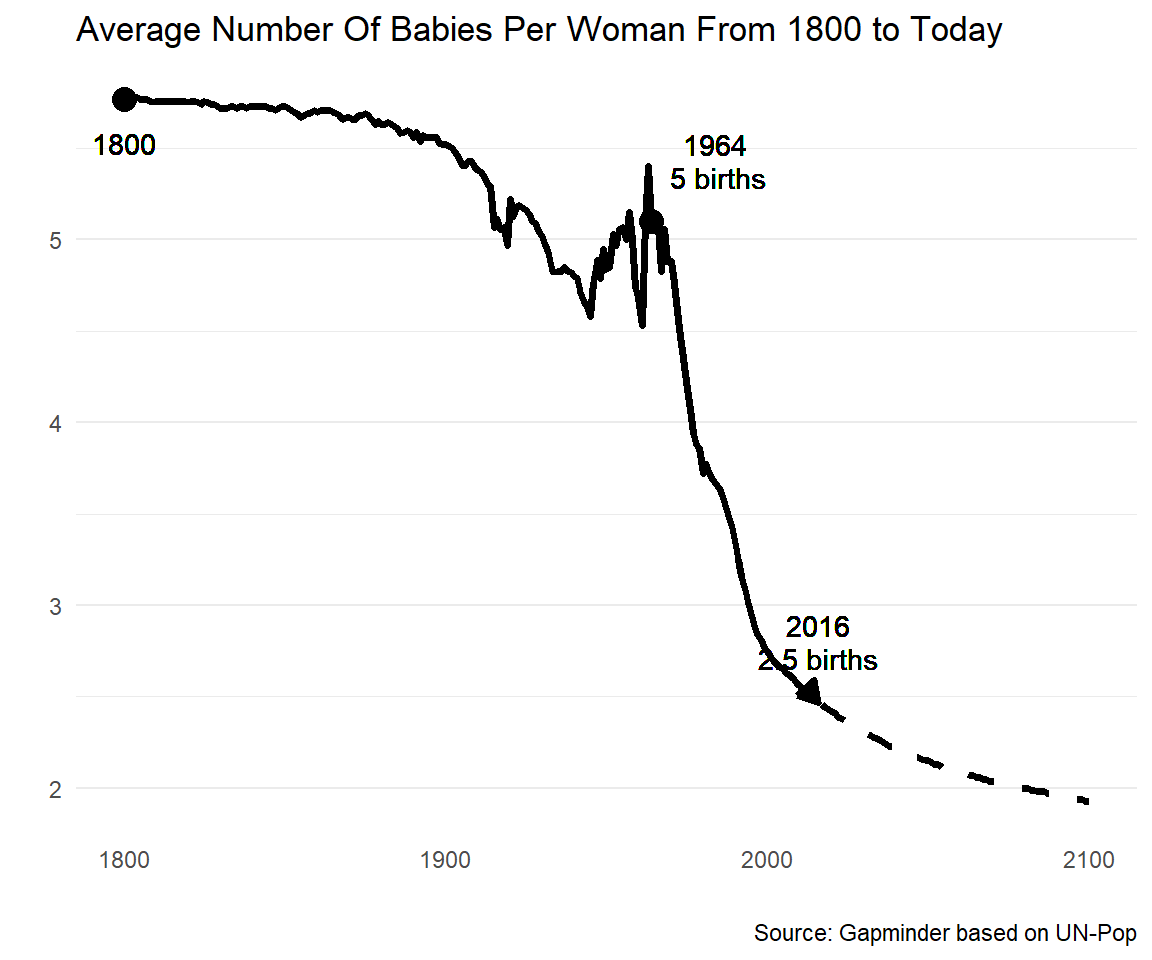
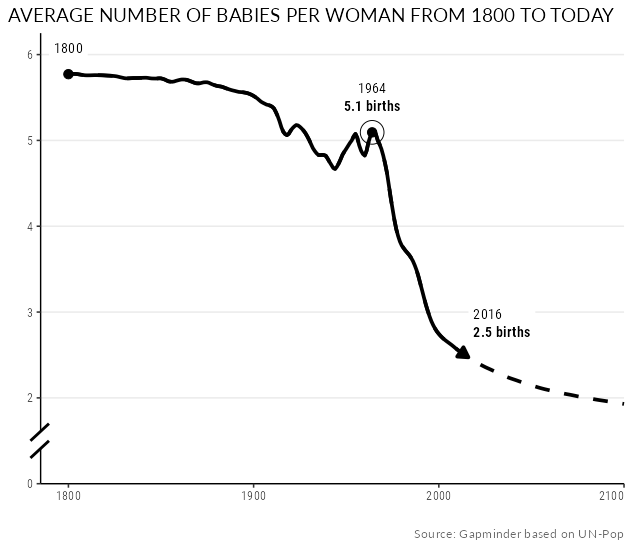
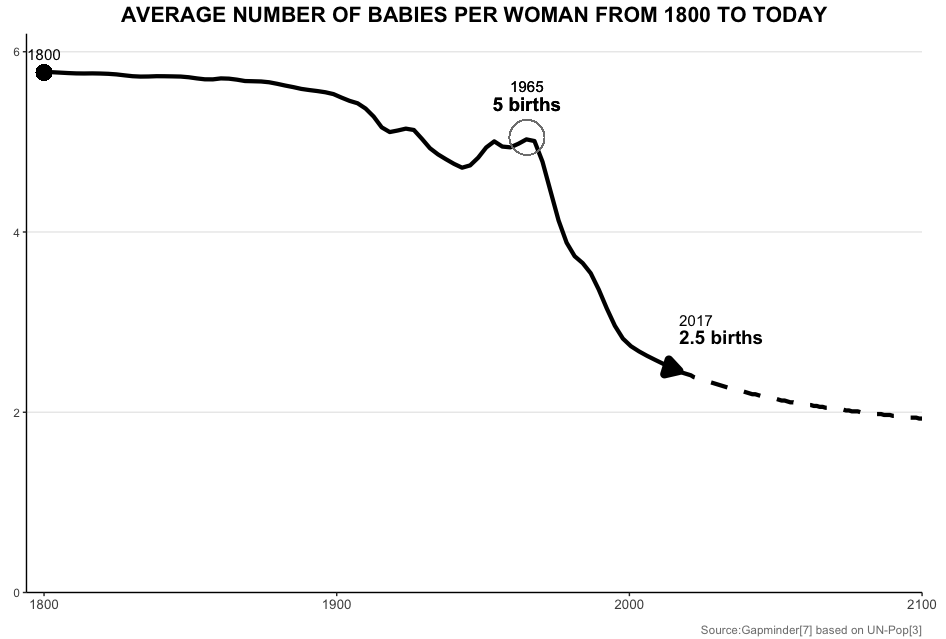
I try to replicate the fertility graph from gapminder as good as I can in ggplot:
I have:
fertility <- read.csv("https://raw.githubusercontent.com/MarcoKuehne/marcokuehne.github.io/main/data/Gapminder/tfr-by-gapminder-v12-20171212.csv", sep = ";")
# manipulate
library(tidyverse)
fertility <- fertility %>%
select(!c(geo.name, geo, indicator)) %>%
rownames_to_column %>%
gather(var, value, -rowname) %>%
spread(rowname, value) %>%
rename(year = var, fert = `1`) %>%
slice_head(n = 301)
fertility[,1:2] <- sapply(fertility[,1:2],FUN=as.numeric)
fertility1 <- fertility[1:217,]
fertility2 <- fertility[218:301,]
# visualize
ggplot(data = fertility, aes(x=year, y=fert))
geom_point(aes(x=year[1], y=fert[1]), size = 4)
geom_text(aes(x=year[1], y=fert[1], label = year[1]), vjust = 2.5)
geom_point(aes(x=year[165], y=fert[165]), size = 4)
geom_text(aes(x=year[165], y=fert[165], label = year[165]), vjust = -3, hjust = -0.5)
geom_text(aes(x=year[165], y=fert[165], label = "5 births"), vjust = -1.5, hjust = -0.2)
#geom_point(aes(x=year[217], y=fert[217]), size = 4)
geom_text(aes(x=year[217], y=fert[217], label = year[217]), vjust = -3)
geom_text(aes(x=year[217], y=fert[217], label = "2.5 births"), vjust = -1.5)
geom_line(data = fertility1, size=1.3, arrow=arrow(length=unit(0.30,"cm"), ends="last", type = "closed"))
geom_line(data = fertility2, size=1.3,linetype="dashed")
labs(title = "Average Number Of Babies Per Woman From 1800 to Today",
caption = "Source: Gapminder based on UN-Pop", y="", x="")
theme_minimal()
theme(panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank())
I have some troubles with the linewidth of geom_line(). And I am looking for a compromise between geom_line() and geom_smooth() to make the line plot just a little bit more smooth.
But my major concern at the moment is the broken y-axis. When y-axis does not start at zero, I'd like to hightlight this as in the gapminder graph.
CodePudding user response:
Following the suggestion by @Roland you could smooth your lines using a loess curve. However, instead of trying with geom_smooth I would suggest to do the calculation outside of ggplot as we need the smoothed values also for the points and the labels and we also need the same values for "both" lines.
For your axis break a simple approach would be to use two annotate, one to place the segments, one to add the filled "gap" for which I use a ribbon. This requires some fiddling to get the right positions, to fix the limits and to set clip="off". Also note that I added the axis lines via geom_h/vline so that the annotate layers could be placed on top of the axes.
Finally, I slightly adjusted your data wrangling code, use a dataset to place the labels and points and instead of using vjust I shift the labels directly via the y position.
EDIT For the final touch I added some additional styling.
# manipulate
library(tidyverse)
library(showtext)
library(hrbrthemes)
font_add_google("lato", "Lato", regular.wt = 900)
fertility <- fertility %>%
select(!c(geo.name, geo, indicator)) %>%
mutate(
across(-indicator.name, as.character),
across(-indicator.name, ~ readr::parse_number(.x, locale = locale(decimal_mark = ",")))
) %>%
pivot_longer(-indicator.name, names_to = "year", values_to = "fert", names_prefix = "X") %>%
mutate(year = as.numeric(year)) |>
mutate(fert_smooth = predict(loess(fert ~ year, span = .05)))
fertility_to_high <- fertility |>
filter(year %in% c(1800, 1964, 2016)) |>
mutate(
label = if_else(!year == 1800, paste0(year, "<br>**", round(fert, 1), " births**"), as.character(year)),
hjust = if_else(year == 2016, 0, .5)
)
ggplot(data = fertility, aes(x = year, y = fert_smooth))
geom_point(data = subset(fertility_to_high, year == 1964), size = 10, shape = 21, fill = NA, color = "black")
geom_point(data = subset(fertility_to_high, year != 2016), size = 3)
ggtext::geom_richtext(data = fertility_to_high, aes(y = fert .15, label = label, hjust = hjust),
vjust = 0, label.colour = NA, family = font_rc, lineheight = 1.2)
geom_line(linewidth = 1.3, linetype = "dashed")
geom_line(data = ~ subset(.x, year <= 2016), linewidth = 1.3, arrow = arrow(length = unit(0.30, "cm"), ends = "last", type = "closed"))
geom_hline(yintercept = 1)
geom_vline(xintercept = 1785)
annotate(geom = "ribbon", x = c(1780, 1790) - .55, ymin = c(1.3, 1.5), ymax = c(1.5, 1.7), fill = "white")
annotate(
geom = "segment",
x = c(1780, 1780) - .5, xend = c(1790, 1790) - .5,
y = c(1.3, 1.5), yend = c(1.5, 1.7), linewidth = 1
)
scale_y_continuous(breaks = 1:6, labels = c(0, 2:6), expand = c(0, .0, .05, 0))
scale_x_continuous(expand = c(0.05, 0, 0, 0))
labs(
title = toupper("Average Number Of Babies Per Woman From 1800 to Today"),
caption = "Source: Gapminder based on UN-Pop", y = "", x = ""
)
coord_cartesian(clip = "off", ylim = c(1, 6), xlim = c(1800, NA))
theme_bw(base_family = font_rc, base_size = 12)
theme(
panel.grid.major.x = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
plot.title = element_text(family = "lato"),
plot.title.position = "plot",
plot.caption = element_text(family = "lato", color = "grey40"),
axis.text.x = element_text(hjust = c(rep(.5, 3), 1))
)
CodePudding user response:
The key is to add:
scale_y_continuous(limits = c(0, 6.2), expand = c(0, 0))
It tells R to limit the y range within 0 and 6.2, and show no gap beyond these two values. The sample code below may work for you, after running it, you will get:
fertility <- read.csv("https://raw.githubusercontent.com/MarcoKuehne/marcokuehne.github.io/main/data/Gapminder/tfr-by-gapminder-v12-20171212.csv", sep = ";")
# manipulate
library(tidyverse)
fertility <- fertility |>
select(!c(geo.name, geo, indicator)) |>
t() |>
as.data.frame() |>
rownames_to_column() |>
slice(-1) |>
as_tibble() |>
rename(c(year = rowname, fert = V1)) |>
mutate(year = str_remove(year, "X"),
year = as.Date(ISOdate(year, 1, 1)),
fert = str_replace(fert, ",", "."),
fert = as.numeric(fert),
fert_1 = case_when(year <= as.Date("2017-01-01") ~ fert,
TRUE ~ as.numeric(NA)),
fert_2 = case_when(year >= as.Date("2017-01-01") ~ fert,
TRUE ~ as.numeric(NA)),
arr_data = case_when((year > as.Date("2014-01-01") & year < "2018-01-01") ~ fert,
TRUE ~ as.numeric(NA)))
fertility |>
ggplot(aes(x = year,
y = fert))
geom_smooth(aes(x = year,
y = fert_1,
group = 1),
span = 0.11,
se = FALSE,
colour = "black",
size = 1.5)
geom_line(aes(x = year,
y = arr_data),
arrow = arrow(length=unit(0.4,"cm"),
ends="last",
type = "closed"),
size = 3)
geom_line(aes(x = year,
y = fert_2,
group = 1),
linetype = 2,
size = 1.5)
geom_point(aes(x = year[1],
y = fert[1]),
size = 5)
annotate(geom = "text",
x = fertility$year[1],
y = fertility$fert[1],
label = "1800",
size = 4,
vjust = -1.2)
geom_point(aes(x = fertility$year[166],
y = fertility$fert[166]),
shape = 1,
size = 12,
colour = "grey50")
annotate(geom = "text",
x = fertility$year[166],
y = fertility$fert[166],
label = "1965",
size = 4,
vjust = -4.2)
annotate(geom = "text",
x = fertility$year[166],
y = fertility$fert[166],
label = "5 births",
size = 5,
fontface = "bold",
vjust = -2)
geom_point(aes(x = fertility$year[166],
y = fertility$fert[166]),
shape = 1,
size = 12,
colour = "grey50")
annotate(geom = "text",
x = fertility$year[166],
y = fertility$fert[166],
label = "1965",
size = 4,
vjust = -4.2)
annotate(geom = "text",
x = fertility$year[166],
y = fertility$fert[166],
label = "5 births",
size = 5,
fontface = "bold",
vjust = -2)
annotate(geom = "text",
x = fertility$year[218],
y = fertility$fert[218],
label = "2017",
size = 4,
vjust = -4.2,
hjust = 0)
annotate(geom = "text",
x = fertility$year[218],
y = fertility$fert[218],
label = "2.5 births",
size = 5,
fontface = "bold",
vjust = -2,
hjust = 0)
theme_bw()
scale_x_date(expand = expansion(mult = c(0.02, 0)))
theme(
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank(),
panel.border = element_blank(),
axis.text.x = element_text(size = 10),
axis.line.x = element_line(),
axis.line.y = element_line(),
plot.caption = element_text(colour = "grey50"),
plot.title = element_text(size = 16,
hjust = 0.5,
face = "bold"),
plot.margin = margin(r = 18,
t = 5,
l = 5,
b = 2)
)
labs(
title = "AVERAGE NUMBER OF BABIES PER WOMAN FROM 1800 TO TODAY",
x = element_blank(),
y = element_blank(),
caption = "Source:Gapminder[7] based on UN-Pop[3]"
)