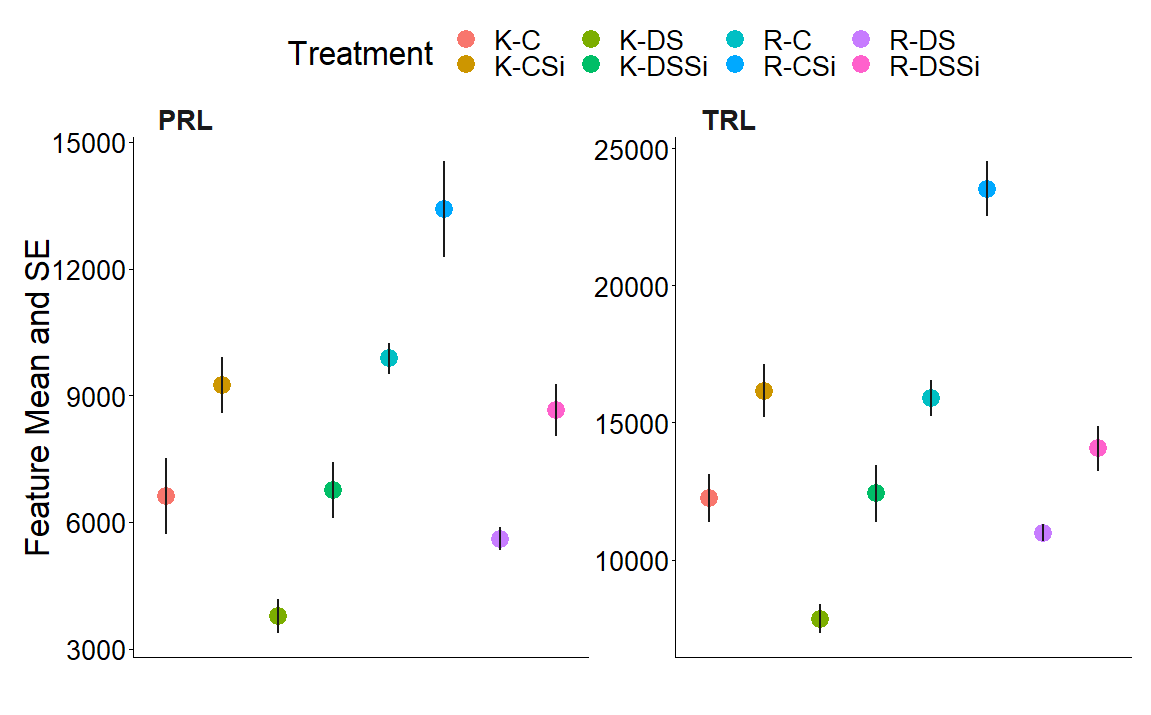How can I calculate p-value from sample mean and se? Thanks for your help.
Below is the code that I am using.
ggplot(combine_mean_se2, aes(x=Treatment, y=mean, colour=Treatment))
geom_point(size=6)
geom_linerange(aes(ymin= mean-se, ymax=mean se), colour="grey10", linewidth=1)
#scale_color_manual(values=c("firebrick4", "dodgerblue4"))
ylab("Feature Mean and SE")
xlab("")
facet_wrap(~traits, scales = "free")
elitetheme2
theme(legend.position = "top",
strip.text.x = element_text(size = 21, face = "bold"))
here is my data
https://docs.google.com/spreadsheets/d/1q1guxAKosKztNXWO3ey1ZKZm4VU49ub58_C_bZ8TO1k/edit#gid=0
CodePudding user response:
You already calculated the mean and se. It would be best if you had each sample value for statistical testing. The ggpubr package has excellent functions to add p-value and SE in your graphs.
http://www.sthda.com/english/wiki/one-way-anova-test-in-r
CodePudding user response:
I may not be correct on this p value, but please let me know if it isn't and let me know any formula to derive the p value
I used the below formula
z = Est/SE
P = exp(−0.717×z − 0.416×z^2)
code
library(rvest)
library(tidyverse)
df <- rvest::read_html('https://docs.google.com/spreadsheets/d/1q1guxAKosKztNXWO3ey1ZKZm4VU49ub58_C_bZ8TO1k/edit#gid=0') %>% html_table()
df2 <- df[[1]][2:17,2:6]
nam <- df[[1]][1,2:6]
nam2 <- (nam %>% pivot_longer(c('A','B','C','D','E')))$value
names(df2) <- nam2
df3 <- df2 %>% mutate(mean=as.numeric(mean), se=as.numeric(se), z=mean/se,
p=exp(-0.717*z-0.416*z^2))
output
# A tibble: 16 × 7
Treatment Si traits mean se z p
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 K-C 0mM TRL 12246. 861. 14.2 1.08e- 41
2 K-CSi 4mM TRL 16164. 979. 16.5 4.38e- 55
3 K-DS 0mM TRL 7852. 525. 14.9 9.67e- 46
4 K-DSSi 4mM TRL 12417. 1039. 12.0 2.94e- 30
5 R-C 0mM TRL 15893. 638. 24.9 1.59e-120
6 R-CSi 4mM TRL 23529. 1001. 23.5 6.29e-108
7 R-DS 0mM TRL 10988. 312. 35.2 5.44e-236
8 R-DSSi 4mM TRL 14063. 818. 17.2 1.70e- 59
9 K-C 0mM PRL 6620. 903. 7.33 9.91e- 13
10 K-CSi 4mM PRL 9237. 662. 14.0 2.96e- 40
11 K-DS 0mM PRL 3779. 408. 9.26 4.16e- 19
12 K-DSSi 4mM PRL 6752. 657. 10.3 5.32e- 23
13 R-C 0mM PRL 9882. 366. 27.0 3.64e-141
14 R-CSi 4mM PRL 13409. 1138. 11.8 1.87e- 29
15 R-DS 0mM PRL 5605. 275. 20.4 2.72e- 82
16 R-DSSi 4mM PRL 8652. 618. 14.0 1.58e- 40