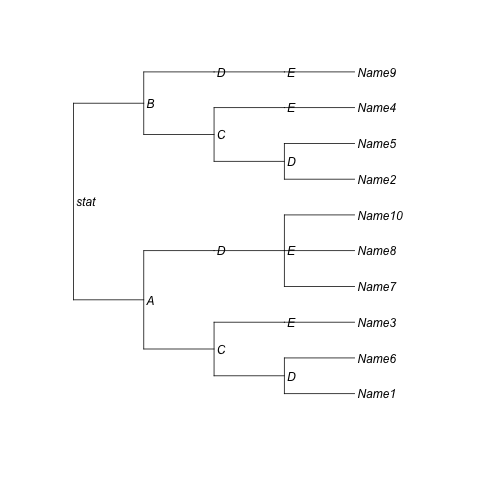
I want to create a graph with the classification of several items (Name1, Name2, Name3, ...) according to several criteria (Column1, Column2, Column3) in the form of a dendrogram in R (I like the orthogonal esthetics of dendrograms to this end).
I have the following CSV (data.csv) with the items and their classification in each criterion:
Name;Column1;Column2;Column3
Name1;A;C;D
Name2;B;C;D
Name3;A;C;E
Name4;B;C;E
Name5;B;C;D
Name6;A;C;D
Name7;A;D;E
Name8;A;D;E
Name9;B;D;E
Name10;A;D;E
And the following R code:
library(data.tree)
library(DiagrammeR)
library(ggdendro)
data <- read.table(file = "data.csv", header = TRUE, sep = ";")
data$pathString <- paste("stat",
data$Column1,
data$Column2,
data$Column3,
data$Name,
sep = "/")
pop <- as.Node(data)
den <- as.dendrogram(pop)
p1 <- ggdendrogram(den, labels = TRUE, rotate = TRUE, leaf_labels = TRUE)
p1
Currently, I'm obtaining the following image without labels in non-leaf nodes:
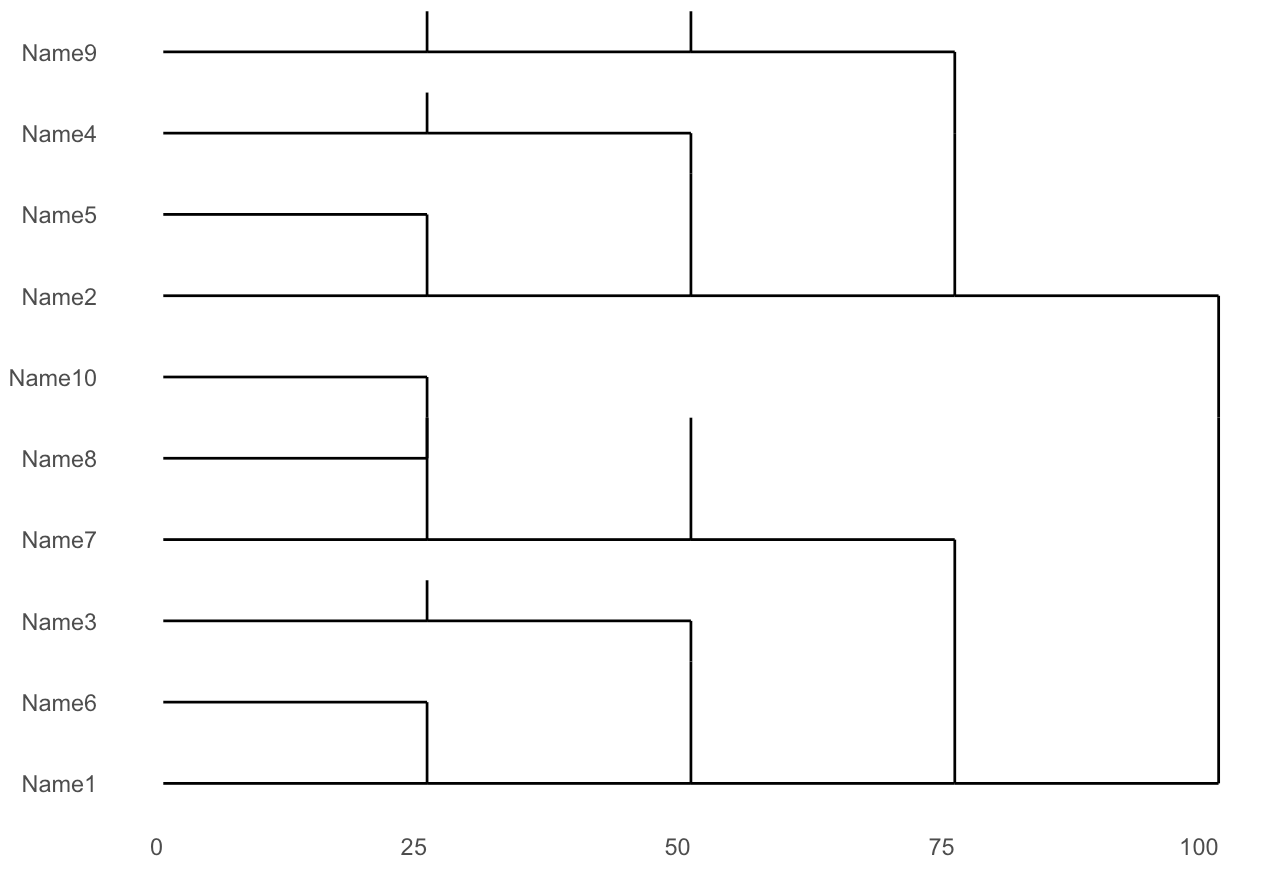
I'm wondering if it is possible to show the labels in each of non-leaf nodes in the dendrogram. For example, show "A" and "B" over each edge (related to non-leaf nodes) in decisions of Column1, show "C" and "D" over each edge in decisions of Column2, and so on. Furthermore, it is possible to put the name of the items at the right hand of the graphic? Thanks in advance.
CodePudding user response:
Try using the ape package.
# try loading a package, install if unavailable
for(i in c("ape")){
if(!require(i, character.only = TRUE)){
install.packages(i, dependencies = TRUE)
library(i, character.only = TRUE)
}
}
# convert "Node" object to class "phylo"
den2 <- as.phylo(pop)
# plot.phylo options allow flexibility in display
plot(den2, show.node.label = TRUE, label.offset = 1)