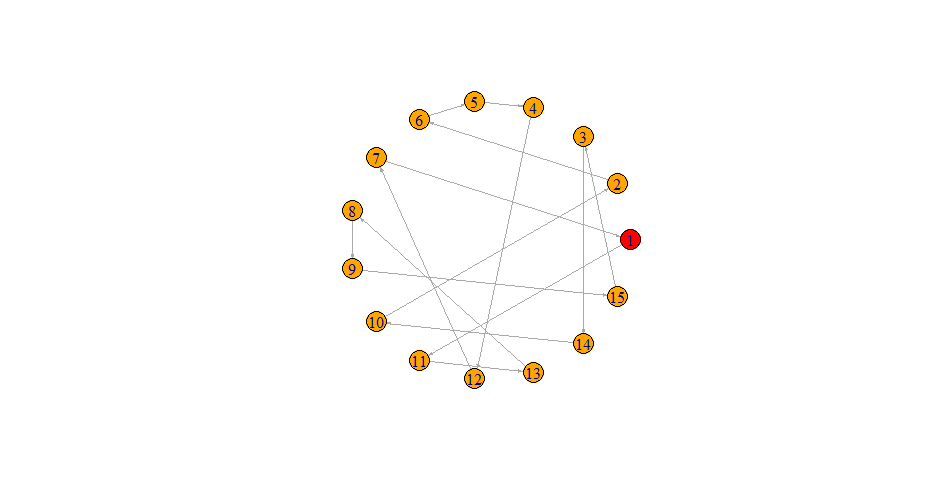
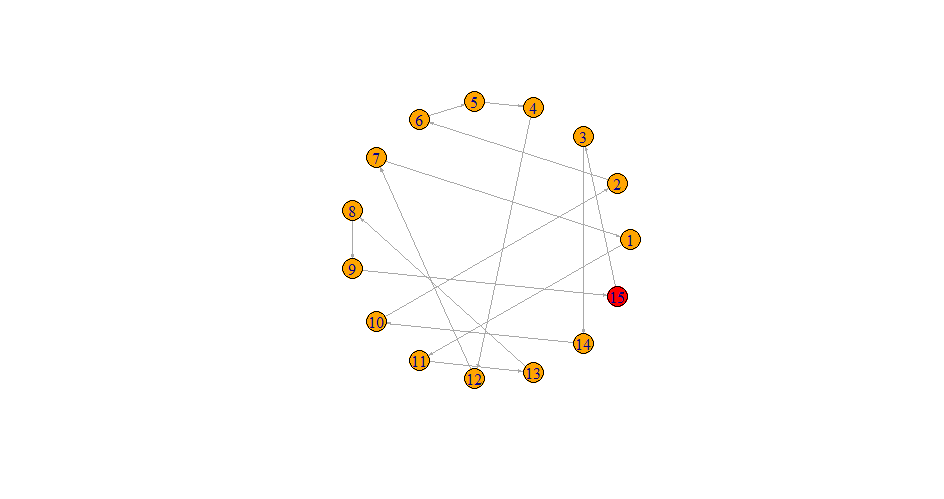
Consider the following points : 1,2,3,4,5,6,7,8,9,10
I can make a directed random graph from these numbers :
library(igraph)
from = sample(1:10, 10, replace=FALSE)
to = sample(1:10, 10, replace=FALSE)
graph_data = data.frame(from,to)
graph_data$id = as.factor(1:10)
graph <- graph.data.frame(graph_data, directed=T)
graph = simplify(graph)
V(graph)$color <- ifelse(graph_data$id == 1, "red", "orange")
plot(graph, layout=layout.circle, edge.arrow.size = 0.2)
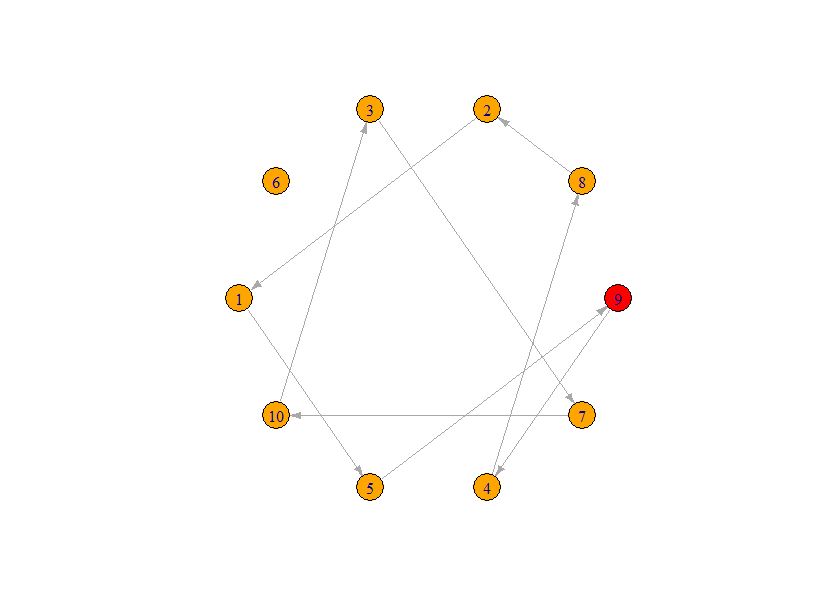
I would like to make changes (i.e. "conditions") to the above graph so that:
- None of the "nodes" are isolated (e.g. node # 6 is "isolated")
- Each node can only have "incoming edge" and an "outgoing edge" (i.e. if you are on some "node" in this graph, if you keep following the arrows in the forward direction, you will eventually visit every other "node" exactly once and finally come back to the "node" you started from)
- The "last node" (i.e. the "node" in the "to" column of the last row) is always connected to the "first node" (i.e. the "node" in the "from" column of the first row)
In this case, this means that :
from to
1 9 4
2 8 2
3 2 1
4 3 7
5 6 6
6 1 5
7 10 3
8 5 9
9 4 8
10 7 10
An additional row would need to be added so that "node 10" has to be connected to "node 9".
I can do this manually (e.g. graph_data[nrow(graph_data) 1,] = c("10","9", "11", "orange)) but can someone please show me how to automatically add all these conditions to the random graph being generated?
Thank you!
CodePudding user response:
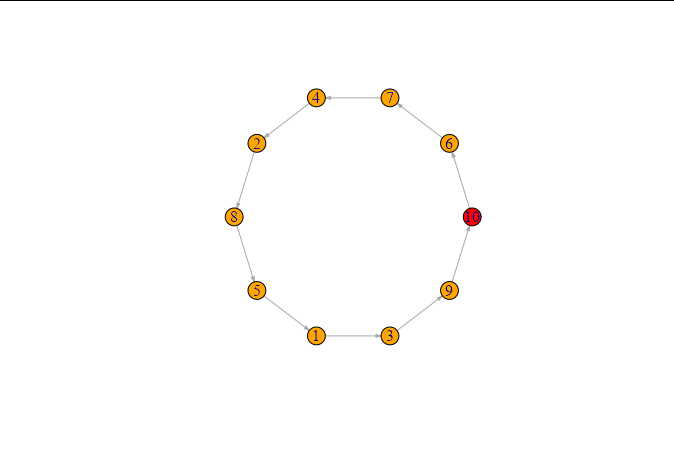
That should solve your problem
library(tidyverse)
library(igraph)
set.seed(123)
n=15
data = tibble(d = paste(1:n))
relations = tibble(
from = sample(data$d),
to = lead(from, default=from[1]),
)
graph = graph_from_data_frame(relations, directed=T, vertices = data)
V(graph)$color <- ifelse(data$d == "1", "red", "orange")
plot(graph, layout=layout.circle, edge.arrow.size = 0.2)
Update 1
library(tidyverse)
library(igraph)
set.seed(123)
n=15
data = tibble(d = paste(1:n))
relations = tibble(
from = sample(data$d),
to = lead(from, default=from[1]),
)
graph = graph_from_data_frame(relations, directed=T, vertices = data)
V(graph)$color <- ifelse(data$d == relations$from[1], "red", "orange")
plot(graph, layout=layout.circle, edge.arrow.size = 0.2)
Here you are
CodePudding user response:
I think the following meets your requirements. If you generate from and to like this:
from <- sample(1:10, 10, replace = FALSE)
to <- from[c(2:10, 1)]
Then repeat the rest of your code, you get:
This produces a cyclical graph where the node labels are random numbers between one and 10.