I have the following dataset, where p1 is the number of plant species indicated by column sp1, and p2 is the number of plant species indicated by column sp2, and so on. I want to create a new variable named Count1 for example, which counts the total number of wheat species in each row and paste it into the new variable Count1. For example in row 9 (ID=7), we have 7 wheat plants in total or in row 7 (ID=5), we don't have any wheat, so it will be 0 in Count1 variable. I would appreciate if you help me to solve this.
plt <- data.frame(ID = c(0:10), p1 = c(1,1,1,8,8,8,8,8,4,4,4),
sp1 = c('wheat', 'wheat', 'wheat', 'barley','barley',
'barley','barley','barley', 'rice','rice','rice'),
p2 = c(0,0,0,2,2,2,2,2,2,2,2),
sp2 = c(0,0,0,'rice', 'rice', 'rice', 'rice', 'wheat',
'wheat', 'wheat','wheat'),
p3 = c(0,0,2,2,2,2, 5,5,5,5,5),
sp3= c(0,0,0, 'rice', 'rice', 'rice', 'wheat', 'wheat',
'wheat', 'wheat', 'wheat'))
I would expect the following table.
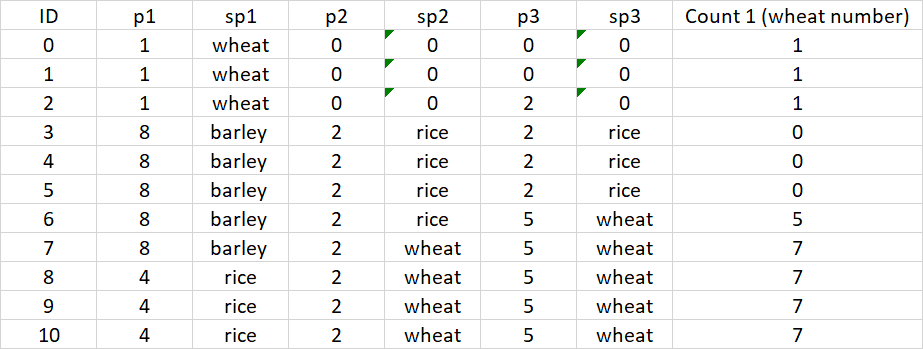
CodePudding user response:
You can use sapply with paste to check your condition and subsequently apply the sum
plt$Count1 <- rowSums(sapply(1:length(grep("^sp",colnames(plt))), function(x)
ifelse(plt[, paste0("sp", x)] == "wheat", plt[, paste0("p", x)], 0)))
plt
ID p1 sp1 p2 sp2 p3 sp3 Count1
1 0 1 wheat 0 0 0 0 1
2 1 1 wheat 0 0 0 0 1
3 2 1 wheat 0 0 2 0 1
4 3 8 barley 2 rice 2 rice 0
5 4 8 barley 2 rice 2 rice 0
6 5 8 barley 2 rice 2 rice 0
7 6 8 barley 2 rice 5 wheat 5
8 7 8 barley 2 wheat 5 wheat 7
9 8 4 rice 2 wheat 5 wheat 7
10 9 4 rice 2 wheat 5 wheat 7
11 10 4 rice 2 wheat 5 wheat 7
CodePudding user response:
With tidyverse you could try the following. First, put your data in long form with pivot_longer, and remove rows where sp is zero.
Then, for each ID and sp, total up the sum of p. You can then put the data into wide form again if desired, and join back to original data. This will calculate sum of each type (wheat, barley, rice).
library(tidyverse)
plt %>%
pivot_longer(cols = -ID, names_to = c(".value", "number"), names_pattern = "(p|sp)(\\d )") %>%
filter(sp != 0) %>%
group_by(ID, sp) %>%
summarise(total = sum(p)) %>%
pivot_wider(id_cols = ID, names_from = sp, values_from = total, values_fill = 0) %>%
right_join(plt)
Output
ID wheat barley rice p1 sp1 p2 sp2 p3 sp3
<int> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <chr>
1 0 1 0 0 1 wheat 0 0 0 0
2 1 1 0 0 1 wheat 0 0 0 0
3 2 1 0 0 1 wheat 0 0 2 0
4 3 0 8 4 8 barley 2 rice 2 rice
5 4 0 8 4 8 barley 2 rice 2 rice
6 5 0 8 4 8 barley 2 rice 2 rice
7 6 5 8 2 8 barley 2 rice 5 wheat
8 7 7 8 0 8 barley 2 wheat 5 wheat
9 8 7 0 4 4 rice 2 wheat 5 wheat
10 9 7 0 4 4 rice 2 wheat 5 wheat
11 10 7 0 4 4 rice 2 wheat 5 wheat
CodePudding user response:
This is a non efficient method, but it's more "intuitive". Ben and Andre's answers are a better solution since its compactness and, probably, its performance.
However, this is the idea:
word <- "wheat" # take class you want
indexes <- which(plt==word,arr.ind = TRUE) # find indices in matrix which contains that word
new_matrix <- subset(plt,is.element(row.names(plt),unique(indexes[,1]))==TRUE) # subset that matrix
count_w <- 0; k <- 1;rowcounts <- c();
for (j in 1:nrow(new_matrix)){ # loop by rows
a <- which(new_matrix[j,]==word) # which columns contains the value "word"?
for (i in a){
count_w <- count_w new_matrix[j,i-1] # Just sum the columns
}
rowcounts[k] <- count_w # save the counts
count_w <- 0; # and start again for another row
k <- k 1
}
Count_1 <- rep(0,nrow(plt));p <- 1
for (k in sort(unique(indexes[,1]))){ # To be consistent with original data frame, new column of counts must have same dimension, so let's fill it with zeros and substitute with counts vector
Count_1[k] <- rowcounts[p]
p <- p 1;
}
final_matrix <- cbind(plt,Count_1) # just combine
You can see that 23 lines of code (in my case) can be substituted with fewer ones (see Ben & Andre's answers)!
