I have data like this:
df<-structure(list(levels_incised___1 = c(0, 0, 0, 0, 0, 0), levels_incised___2 = c(1,
0, 0, 0, 0, 0), levels_incised___3 = c(1, 0, 0, 0, 0, 0), levels_incised___4 = c(1,
0, 0, 0, 0, 0), levels_incised___5 = c(1, 0, 0, 0, 0, 0), levels_incised___6 = c(1,
0, 0, 0, 0, 0), levels_incised___7 = c(1, 0, 0, 0, 0, 0), levels_incised___8 = c(1,
1, 1, 0, 0, 0), levels_incised___9 = c(1, 1, 1, 0, 0, 0), levels_incised___10 = c(1,
1, 1, 0, 0, 0), levels_incised___11 = c(0, 1, 0, 0, 0, 0), levels_incised___12 = c(0,
1, 0, 0, 0, 0), levels_incised___13 = c(0, 1, 0, 0, 0, 0), levels_incised___14 = c(0,
1, 0, 0, 0, 0), levels_incised___15 = c(0, 1, 0, 0, 0, 0), levels_incised___16 = c(0,
0, 0, 0, 0, 0), levels_incised___17 = c(0, 0, 0, 0, 0, 0), levels_incised___18 = c(0,
0, 0, 0, 0, 0), levels_incised___19 = c(0, 0, 0, 0, 0, 0), levels_incised___20 = c(0,
0, 0, 0, 0, 0), levels_incised___21 = c(0, 0, 0, 0, 0, 0), levels_incised___22 = c(0,
0, 0, 0, 1, 0), levels_incised___23 = c(0, 0, 0, 0, 1, 1), levels_incised___24 = c(0,
0, 0, 0, 1, 1), levels_incised___25 = c(0, 0, 0, 0, 1, 1), levels_incised___26 = c(0,
0, 0, 0, 1, 1), levels_incised___27 = c(0, 0, 0, 1, 1, 1), levels_incised___28 = c(0,
0, 0, 1, 1, 1), levels_incised___29 = c(0, 0, 0, 1, 1, 0), levels_incised___30 = c(0,
0, 0, 1, 1, 0), levels_incised___31 = c(0, 0, 0, 0, 0, 0)), row.names = c(NA,
-6L), class = c("tbl_df", "tbl", "data.frame"))
That originally came from this Redcap input where each button was one of those columns:
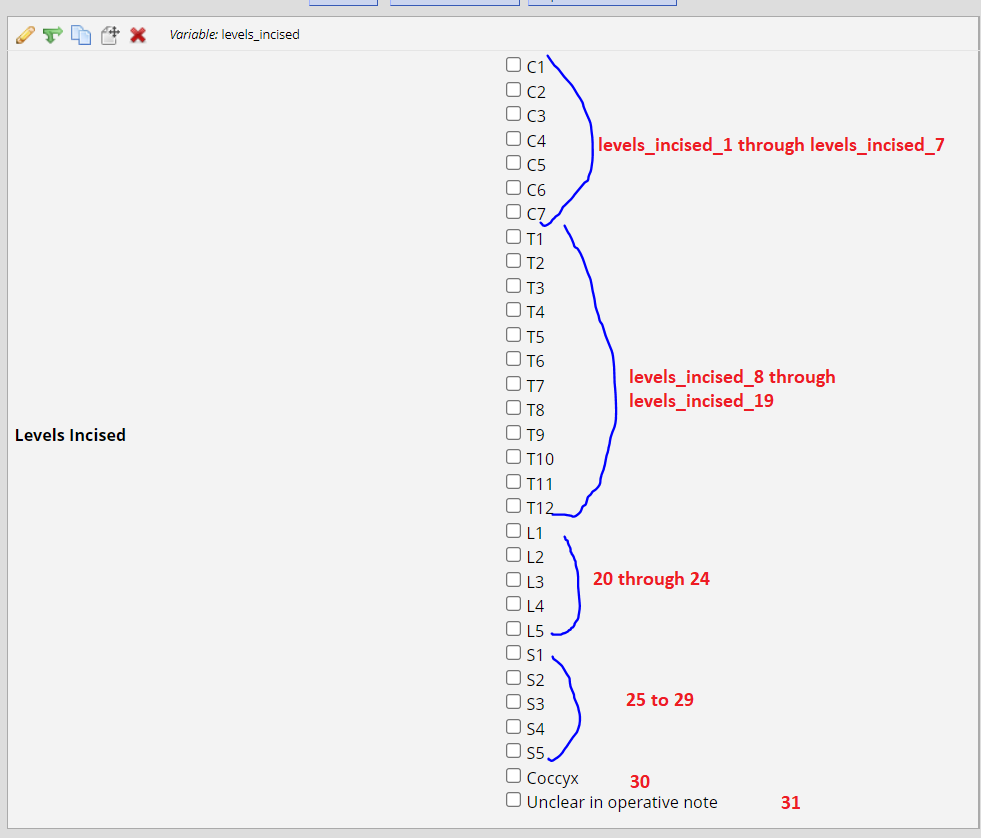
And I need to create a column at the end (lets call it Level) with these possible inputs:
- Cervical (any of the c buttons)
- Thoracic (the t's)
- Lumbar (the L's)
- Sacral (sacral)
- Thoracocervical (t's or c's)
- Thoracolumbar (t's or l's)
- Lumbosacral (l's and sacral)
So for instance, the patient in the first row had "1"'s in levels_incised_2 through levels_incised_10... meaning they had values in both the cervical range and the thoracic range. So that patient should get "Thoracocervical".
The patient in row 2 had 1's in 8 through 15, so they'd only get a "thoracic"
Does anyone know the most straight forward way to accomplish this?
Oh one last detail, there's 100 other columns so it'd be nice if I could select/name these specific ones to count
CodePudding user response:
A few things to resolve here:
- find a way to convert
levels...#to one of theC/T/... categories; - produce logic to infer based on presence of groups.
I think the first can be done by extracting the number and using findInterval to determine with of C/T/... each column belongs to. From there, we can do some simple c_across to find "any" in a group, and case_when to get your Level labels.
library(dplyr)
# helper function for renaming
func <- function(z) {
num <- as.integer(gsub("\\D", "", z))
grp <- c("C","T","L","S","Co","unclear")[findInterval(num, 1 c(0, 7, 19, 24, 29, 30, 31))]
grp <- paste0(grp, ave(grp, grp, FUN = seq_along))
# fix those that do not need numbering
grp[grepl("^Co", grp)] <- "Co"
grp[grepl("^unc", grp)] <- "unclear"
grp
}
out <- df %>%
rename_with(.cols = starts_with("levels"), .fn = func) %>%
rowwise() %>%
mutate(
anyC = sum(c_across(C1:C7)) > 0,
anyT = sum(c_across(T1:T12)) > 0,
anyL = sum(c_across(L1:L5)) > 0,
anyS = sum(c_across(S1:S5)) > 0
) %>%
ungroup() %>%
mutate(
Level = case_when(
anyC & anyT & anyL ~ "More than 2?",
anyL & anyS ~ "Lumbosacral",
anyT & anyL ~ "Thoracolumbar",
anyT & anyC ~ "Thoracocervical",
anyS ~ "Sacral",
anyL ~ "Lumbar",
anyT ~ "Thoracic",
anyC ~ "Cervical",
TRUE ~ "Nothing?"
)
)
out
# # A tibble: 6 x 36
# C1 C2 C3 C4 C5 C6 C7 T1 T2 T3 T4 T5 T6 T7 T8 T9 T10 T11 T12 L1 L2 L3 L4 L5 S1 S2 S3 S4 S5 Co unclear anyC anyT anyL anyS Level
# <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <lgl> <lgl> <lgl> <lgl> <chr>
# 1 0 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 TRUE TRUE FALSE FALSE Thoracocervical
# 2 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 FALSE TRUE FALSE FALSE Thoracic
# 3 0 0 0 0 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 FALSE TRUE FALSE FALSE Thoracic
# 4 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 FALSE FALSE FALSE TRUE Sacral
# 5 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 0 FALSE FALSE TRUE TRUE Lumbosacral
# 6 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 0 0 0 FALSE FALSE TRUE TRUE Lumbosacral
out$Level
# [1] "Thoracocervical" "Thoracic" "Thoracic" "Sacral" "Lumbosacral" "Lumbosacral"
If you don't want to keep the renaming, then you can combine the Level result to your original frame with cbind(df, Level = out$Level).
CodePudding user response:
using package dplyr:
## vertebra codes needed later on
vertebra_codes <- c(
paste0('C',1:7), paste0('T',1:12),
paste0('L',1:5), paste0('S',1:5),
'X', ## for Coccyx
'-' ## for unknown
)
df %>%
mutate(
## assuming each row is a case:
case_id = paste0('case_',row_number())
) %>%
## reshape the data from wide to long format:
pivot_longer(
cols = -case_id,
names_to = 'level_incised', values_to = 'is_incised'
) %>%
mutate(
## remove the redundant 'levels_incised__' prefix:
level_incised = gsub('.*_','',level_incised),
## assign the vertebra corresponding to 'level':
vertebra = vertebra_codes[as.integer(level_incised)],
## assign the spine region (e.g.: all lumbal vert. start with 'L'
spine_region = substr(vertebra,1,1)
) %>%
filter(is_incised == 1) %>% ## we're interested in incised vert. only
## remove replicates (more than one vertebra per spine region affected:
distinct(case_id, spine_region) %>%
## do the counts per case:
group_by(case_id) %>%
## string together the affected regions per case:
summarise(incised_regions = paste(spine_region, collapse = ','))
result:
# A tibble: 6 x 2
case_id incised_regions
<chr> <chr>
1 case_1 C,T
2 case_2 T
3 case_3 T
4 case_4 S,X
5 case_5 L,S,X
6 case_6 L,S
(Note that original `df` remains unchanged throughout the processing pipeline. However you can break up the pipeline by removing the `%>%` operator and inspect the intermediary steps, or assign them to temporary objects.)
extra / for fun: example code to ggplot the spine with vertebra status (incised or not) per patient.
