I am working with the R programming language.
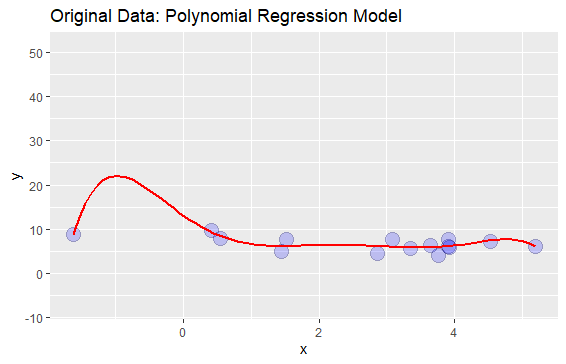
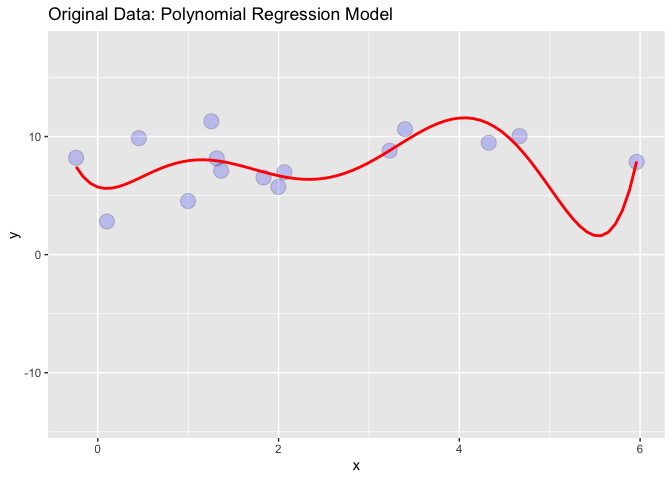
I generated some random data and added a polynomial regression line to the data:
# PLOT 1
library(ggplot2)
x = rnorm(15, 2,2)
y = rnorm(15,7,2)
df = data.frame(x,y)
p <-ggplot(df, aes(x, y))
p <- p geom_point(alpha=2/10, shape=21, fill="blue", colour="black", size=5)
#Add a loess smoother
p stat_smooth(method="lm", se=TRUE, fill=NA, formula=y ~ poly(x, 6, raw=TRUE),colour="red") ggtitle("Original Data: Polynomial Regression Model")
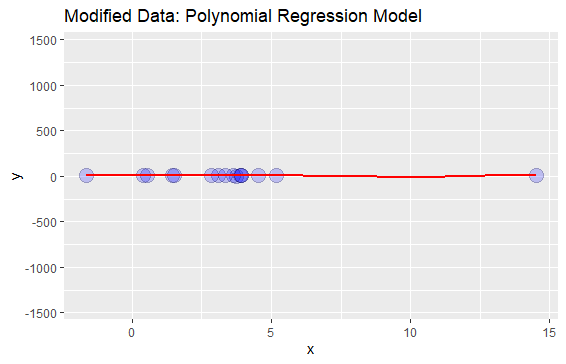
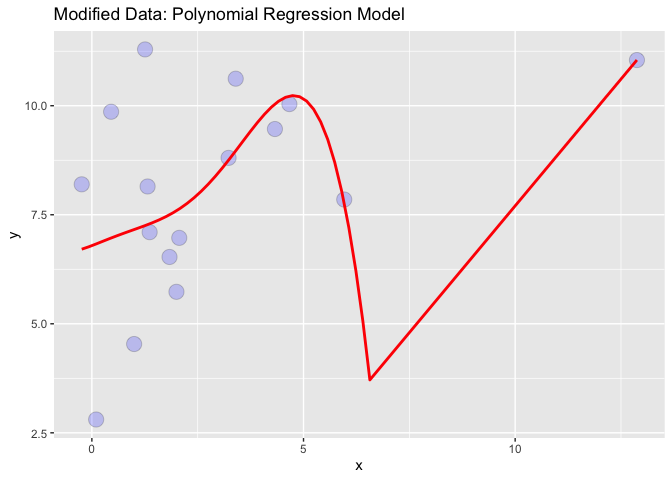
Now, I want to add a single outlier to this data, re-fit the polynomial regression and plot the data:
# PLOT 2
x = rnorm(1,13,1)
y = rnorm(1, 13,1)
df_1 = data.frame(x,y)
df = rbind(df, df_1)
p <-ggplot(df, aes(x, y))
p <- p geom_point(alpha=2/10, shape=21, fill="blue", colour="black", size=5)
#Add a loess smoother
p stat_smooth(method="lm", se=TRUE, fill=NA,
formula=y ~ poly(x, 6, raw=TRUE),colour="red") ggtitle("Modified Data: Polynomial Regression Model")
My Problem: The problem is, now the axis has become so big that the data looks like a "flat line":
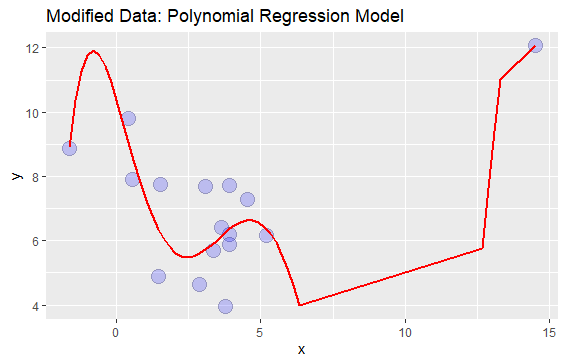
I tried to fix this by limiting the size of the axis:
p stat_smooth(method="lm", se=TRUE, fill=NA, formula=y ~ poly(x, 6, raw=TRUE),colour="red") ggtitle("Modified Data: Polynomial Regression Model") scale_y_continuous(limits = c(min(df$y),max(df$y)))
But I now get the following warning message:
Warning message:
Removed 35 rows containing missing values (geom_smooth).
My Question: Why are rows being deleted when I try to fix the axis? Is there a better way to correct this problem?
Thanks!
CodePudding user response:
When you fit a stat_smooth() (or geom_smooth()) curve you are essentially creating data points i.e. you are generating a list of coordinates that the line will follow. When you changed the y axis limits, some of these coordinates ended up outside the limits and were removed. So, it isn't your original 16 points that are outside your limits, it is the 'calculated' coordinates for the geom_smooth() line.
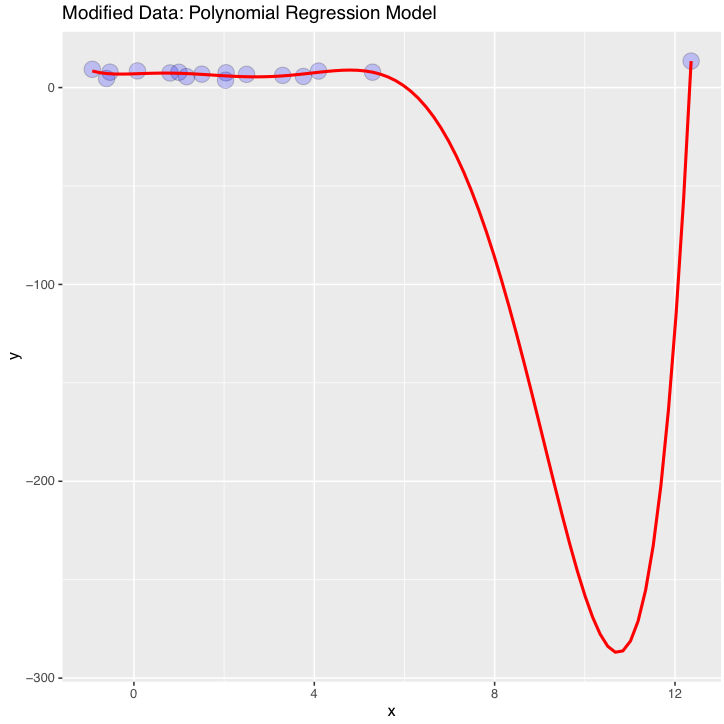
Here is an example showing the new 'internal' data created by stat_smooth() in the ggplot object ("p2"):
library(ggplot2)
x = rnorm(15, 2,2)
y = rnorm(15,7,2)
df = data.frame(x,y)
p <- ggplot(df, aes(x, y))
geom_point(alpha=2/10, shape=21,
fill="blue", colour="black",
size=5)
geom_smooth(method="lm", se=TRUE, fill=NA,
formula=y ~ poly(x, 6, raw=TRUE),
colour="red")
ggtitle("Original Data: Polynomial Regression Model")
p
x = rnorm(1,13,1)
y = rnorm(1, 13,1)
df_1 = data.frame(x,y)
df = rbind(df, df_1)
p2 <-ggplot(df, aes(x, y))
geom_point(alpha=2/10, shape=21,
fill="blue", colour="black",
size=5)
stat_smooth(method="lm", se=TRUE, fill=NA,
formula=y ~ poly(x, 6, raw=TRUE),
colour="red")
ggtitle("Modified Data: Polynomial Regression Model")
scale_y_continuous(limits = c(min(df$y),max(df$y)))
p2
#> Warning: Removed 37 rows containing missing values (geom_smooth).
ggplot_build(p2)$data[[2]]
#> x y ymin ymax se flipped_aes PANEL group
#> 1 -0.23985422 6.710141 NA NA 2.432118 FALSE 1 -1
#> 2 -0.07381912 6.765093 2.975488 10.554698 1.675217 FALSE 1 -1
#> 3 0.09221597 6.827052 3.150370 10.503735 1.625299 FALSE 1 -1
#> 4 0.25825106 6.891454 3.020561 10.762347 1.711151 FALSE 1 -1
#> 5 0.42428615 6.955375 3.107609 10.803142 1.700928 FALSE 1 -1
#> 6 0.59032125 7.017292 3.429403 10.605180 1.586047 FALSE 1 -1
#> 7 0.75635634 7.076844 3.875816 10.277873 1.415034 FALSE 1 -1
#> 8 0.92239143 7.134617 4.321137 9.948098 1.243716 FALSE 1 -1
#> 9 1.08842652 7.191933 4.663619 9.720246 1.117656 FALSE 1 -1
#> 10 1.25446161 7.250651 4.856861 9.644442 1.058189 FALSE 1 -1
#> 11 1.42049671 7.312991 4.923653 9.702330 1.056221 FALSE 1 -1
#> 12 1.58653180 7.381355 4.924840 9.837869 1.085917 FALSE 1 -1
#> 13 1.75256689 7.458168 4.912355 10.003980 1.125392 FALSE 1 -1
#> 14 1.91860198 7.545735 4.908758 10.182712 1.165691 FALSE 1 -1
#> 15 2.08463707 7.646104 4.911585 10.380623 1.208810 FALSE 1 -1
#> 16 2.25067217 7.760941 4.907110 10.614772 1.261553 FALSE 1 -1
#> 17 2.41670726 7.891421 4.884922 10.897920 1.329041 FALSE 1 -1
#> 18 2.58274235 8.038130 4.847749 11.228511 1.410327 FALSE 1 -1
#> 19 2.74877744 8.200977 4.813006 NA 1.497673 FALSE 1 -1
#> 20 2.91481253 8.379121 4.807386 NA 1.578907 FALSE 1 -1
#> 21 3.08084763 8.570910 4.858931 NA 1.640902 FALSE 1 -1
#> 22 3.24688272 8.773830 4.989796 NA 1.672755 FALSE 1 -1
#> 23 3.41291781 8.984466 5.210253 NA 1.668413 FALSE 1 -1
#> 24 3.57895290 9.198480 5.512992 NA 1.629192 FALSE 1 -1
#> 25 3.74498800 9.410595 5.866693 NA 1.566603 FALSE 1 -1
#> 26 3.91102309 9.614597 6.209727 NA 1.505143 FALSE 1 -1
#> 27 4.07705818 9.803344 6.451282 NA 1.481799 FALSE 1 -1
#> 28 4.24309327 9.968788 6.496294 NA 1.535037 FALSE 1 -1
#> 29 4.40912836 10.102016 6.295586 NA 1.682655 FALSE 1 -1
#> 30 4.57516346 10.193293 5.875238 NA 1.908822 FALSE 1 -1
#> 31 4.74119855 10.232124 5.314615 NA 2.173814 FALSE 1 -1
#> 32 4.90723364 10.207325 4.710764 NA 2.429787 FALSE 1 -1
#> 33 5.07326873 10.107110 4.159409 NA 2.629216 FALSE 1 -1
#> 34 5.23930382 9.919186 3.745953 NA 2.728914 FALSE 1 -1
#> 35 5.40533892 9.630862 3.530453 NA 2.696722 FALSE 1 -1
#> 36 5.57137401 9.229169 3.502837 NA 2.531359 FALSE 1 -1
#> 37 5.73740910 8.700996 3.447417 NA 2.322376 FALSE 1 -1
#> 38 5.90344419 8.033234 NA NA 2.378236 FALSE 1 -1
#> 39 6.06947929 7.212932 NA NA 3.159180 FALSE 1 -1
#> 40 6.23551438 6.227470 NA NA 4.805205 FALSE 1 -1
#> 41 6.40154947 5.064739 NA NA 7.234080 FALSE 1 -1
#> 42 6.56758456 3.713337 NA NA 10.414227 FALSE 1 -1
#> 43 6.73361965 NA NA NA 14.368943 FALSE 1 -1
#> 44 6.89965475 NA NA NA 19.144526 FALSE 1 -1
#> 45 7.06568984 NA NA NA 24.794757 FALSE 1 -1
#> 46 7.23172493 NA NA NA 31.373858 FALSE 1 -1
#> 47 7.39776002 NA NA NA 38.932662 FALSE 1 -1
#> 48 7.56379511 NA NA NA 47.516027 FALSE 1 -1
#> 49 7.72983021 NA NA NA 57.160715 FALSE 1 -1
#> 50 7.89586530 NA NA NA 67.893447 FALSE 1 -1
#> 51 8.06190039 NA NA NA 79.728997 FALSE 1 -1
#> 52 8.22793548 NA NA NA 92.668271 FALSE 1 -1
#> 53 8.39397057 NA NA NA 106.696330 FALSE 1 -1
#> 54 8.56000567 NA NA NA 121.780347 FALSE 1 -1
#> 55 8.72604076 NA NA NA 137.867491 FALSE 1 -1
#> 56 8.89207585 NA NA NA 154.882727 FALSE 1 -1
#> 57 9.05811094 NA NA NA 172.726539 FALSE 1 -1
#> 58 9.22414604 NA NA NA 191.272557 FALSE 1 -1
#> 59 9.39018113 NA NA NA 210.365110 FALSE 1 -1
#> 60 9.55621622 NA NA NA 229.816685 FALSE 1 -1
#> 61 9.72225131 NA NA NA 249.405299 FALSE 1 -1
#> 62 9.88828640 NA NA NA 268.871784 FALSE 1 -1
#> 63 10.05432150 NA NA NA 287.916987 FALSE 1 -1
#> 64 10.22035659 NA NA NA 306.198879 FALSE 1 -1
#> 65 10.38639168 NA NA NA 323.329573 FALSE 1 -1
#> 66 10.55242677 NA NA NA 338.872263 FALSE 1 -1
#> 67 10.71846186 NA NA NA 352.338062 FALSE 1 -1
#> 68 10.88449696 NA NA NA 363.182766 FALSE 1 -1
#> 69 11.05053205 NA NA NA 370.803517 FALSE 1 -1
#> 70 11.21656714 NA NA NA 374.535386 FALSE 1 -1
#> 71 11.38260223 NA NA NA 373.647868 FALSE 1 -1
#> 72 11.54863733 NA NA NA 367.341284 FALSE 1 -1
#> 73 11.71467242 NA NA NA 354.743103 FALSE 1 -1
#> 74 11.88070751 NA NA NA 334.904184 FALSE 1 -1
#> 75 12.04674260 NA NA NA 306.794933 FALSE 1 -1
#> 76 12.21277769 NA NA NA 269.301428 FALSE 1 -1
#> 77 12.37881279 NA NA NA 221.221573 FALSE 1 -1
#> 78 12.54484788 NA NA NA 161.261789 FALSE 1 -1
#> 79 12.71088297 NA NA NA 88.038458 FALSE 1 -1
#> 80 12.87691806 11.047284 5.267979 NA 2.554776 FALSE 1 -1
#> colour fill size linetype weight alpha
#> 1 red NA 1 1 1 0.4
#> 2 red NA 1 1 1 0.4
#> 3 red NA 1 1 1 0.4
#> 4 red NA 1 1 1 0.4
#> 5 red NA 1 1 1 0.4
#> 6 red NA 1 1 1 0.4
#> 7 red NA 1 1 1 0.4
#> 8 red NA 1 1 1 0.4
#> 9 red NA 1 1 1 0.4
#> 10 red NA 1 1 1 0.4
#> 11 red NA 1 1 1 0.4
#> 12 red NA 1 1 1 0.4
#> 13 red NA 1 1 1 0.4
#> 14 red NA 1 1 1 0.4
#> 15 red NA 1 1 1 0.4
#> 16 red NA 1 1 1 0.4
#> 17 red NA 1 1 1 0.4
#> 18 red NA 1 1 1 0.4
#> 19 red NA 1 1 1 0.4
#> 20 red NA 1 1 1 0.4
#> 21 red NA 1 1 1 0.4
#> 22 red NA 1 1 1 0.4
#> 23 red NA 1 1 1 0.4
#> 24 red NA 1 1 1 0.4
#> 25 red NA 1 1 1 0.4
#> 26 red NA 1 1 1 0.4
#> 27 red NA 1 1 1 0.4
#> 28 red NA 1 1 1 0.4
#> 29 red NA 1 1 1 0.4
#> 30 red NA 1 1 1 0.4
#> 31 red NA 1 1 1 0.4
#> 32 red NA 1 1 1 0.4
#> 33 red NA 1 1 1 0.4
#> 34 red NA 1 1 1 0.4
#> 35 red NA 1 1 1 0.4
#> 36 red NA 1 1 1 0.4
#> 37 red NA 1 1 1 0.4
#> 38 red NA 1 1 1 0.4
#> 39 red NA 1 1 1 0.4
#> 40 red NA 1 1 1 0.4
#> 41 red NA 1 1 1 0.4
#> 42 red NA 1 1 1 0.4
#> 43 red NA 1 1 1 0.4
#> 44 red NA 1 1 1 0.4
#> 45 red NA 1 1 1 0.4
#> 46 red NA 1 1 1 0.4
#> 47 red NA 1 1 1 0.4
#> 48 red NA 1 1 1 0.4
#> 49 red NA 1 1 1 0.4
#> 50 red NA 1 1 1 0.4
#> 51 red NA 1 1 1 0.4
#> 52 red NA 1 1 1 0.4
#> 53 red NA 1 1 1 0.4
#> 54 red NA 1 1 1 0.4
#> 55 red NA 1 1 1 0.4
#> 56 red NA 1 1 1 0.4
#> 57 red NA 1 1 1 0.4
#> 58 red NA 1 1 1 0.4
#> 59 red NA 1 1 1 0.4
#> 60 red NA 1 1 1 0.4
#> 61 red NA 1 1 1 0.4
#> 62 red NA 1 1 1 0.4
#> 63 red NA 1 1 1 0.4
#> 64 red NA 1 1 1 0.4
#> 65 red NA 1 1 1 0.4
#> 66 red NA 1 1 1 0.4
#> 67 red NA 1 1 1 0.4
#> 68 red NA 1 1 1 0.4
#> 69 red NA 1 1 1 0.4
#> 70 red NA 1 1 1 0.4
#> 71 red NA 1 1 1 0.4
#> 72 red NA 1 1 1 0.4
#> 73 red NA 1 1 1 0.4
#> 74 red NA 1 1 1 0.4
#> 75 red NA 1 1 1 0.4
#> 76 red NA 1 1 1 0.4
#> 77 red NA 1 1 1 0.4
#> 78 red NA 1 1 1 0.4
#> 79 red NA 1 1 1 0.4
#> 80 red NA 1 1 1 0.4