I'm currently working on one project where I need to quantize the image. First, I'm reading the image using skimage, and the shape of it is (825, 1100, 3). Image array looks like this:
[[[ 43 78 48]
[ 43 78 48]
[ 43 78 48]
...
[ 5 24 18]
[ 5 24 18]
[ 4 23 17]]
[[ 43 78 48]
[ 43 78 48]
[ 43 78 48]
...
[ 5 24 18]
[ 5 24 18]
[ 4 23 17]]
[[ 43 78 48]
[ 43 78 48]
[ 43 78 48]
...
[ 5 24 18]
[ 4 23 17]
[ 4 23 17]]
...
[[ 99 143 45]
[ 99 143 45]
[ 98 142 44]
...
[102 145 38]
[100 146 38]
[100 146 38]]
[[ 99 143 45]
[ 99 143 45]
[ 99 143 45]
...
[103 146 39]
[100 146 38]
[ 99 145 37]]
[[ 97 142 41]
[ 98 143 42]
[ 99 144 43]
...
[100 146 38]
[ 99 145 37]
[ 99 145 37]]]
Then I apply K-means to quantize the image and decrease the colors in it, and I call that arrary less_colors which also has the same shape of (825, 1100, 3). The output is:
[[[ 29 48 30]
[ 29 48 30]
[ 29 48 30]
...
[ 29 48 30]
[ 29 48 30]
[ 29 48 30]]
[[ 29 48 30]
[ 29 48 30]
[ 29 48 30]
...
[ 29 48 30]
[ 29 48 30]
[ 29 48 30]]
[[ 29 48 30]
[ 29 48 30]
[ 29 48 30]
...
[ 29 48 30]
[ 29 48 30]
[ 29 48 30]]
...
[[111 137 58]
[111 137 58]
[111 137 58]
...
[111 137 58]
[111 137 58]
[111 137 58]]
[[111 137 58]
[111 137 58]
[111 137 58]
...
[111 137 58]
[111 137 58]
[111 137 58]]
[[111 137 58]
[111 137 58]
[111 137 58]
...
[111 137 58]
[111 137 58]
[111 137 58]]]
I have another variable called first which is a list that is [30, 48, 29].
I would like to change the row of less_colors array into a different array (let's say [0, 0, 0]) if it contains the array called first.
I have tried NumPy, but my code does not work.
less_colors[np.where((less_colors == first).all(axis=2))] = [0,0,0]
The complete code:
import cv2
img = io.imread('dog.jpg')
less_colors[(less_colors[:, :] == first).all(axis=2)] = [0, 0, 0]
io.imshow(less_colors)
plt.show()
CodePudding user response:
Short answer:
This was already answered in comments, however, here goes the complete answer:
less_color[(less_color==first).all(axis=2)] = 0
What's goning on?
less_color==first returns a boolean mask which is True only for the indexes where the condition is met. This is a matrix with the same shape as the image.
Next, the .all(axis=2) operation make sure that the condition is met for all the channels (the second axis): you want to overwrite iff three channels contain same value. This also returns a boolean mask, but now with only two dimensions, telling if each coordinate [i,j] accomplish the criteria at the three channels.
Then, we are using this mask to select only those pixels in the less_colors array: less_color[(less_color==first).all(axis=2)]
Finally, we assign those pixels with the desired value, overriding them with 0; note that this is equivalent to [0, 0, 0] due to numpy's broadcasting mechanism.
Small working example
import numpy as np
# create a small image with ones
less_color = np.ones((5,5,3))
# change one pixel with a different value
less_color[1,1] = 30, 40, 29
# This other should kep as is, since only 2 out of three match the required value
less_color[2,2] = 30, 40, 290
print(less_color)
print('='*10)
# the following line actually solves the question
less_color[(less_color==[30, 40, 29]).all(axis=2)] = 0
# check it out:
print(less_color)
Common error:
less_color[less_color==first] = 0 is not enough since it will also replace pixels with partial-matching, for instance, pixels with values like [10, 10, 29] will end up as [10, 10, 0] while they must not be changed.
Thanks @Aaron for your original and quickly answer.
CodePudding user response:
So you want to map a new value to an old value. For your very case it is:
arr[np.all(arr == old_value, axis=-1)] = new_value
But you can create a general function to apply any mapping to any ndarray as follows:
def ndarray_apply_mapping(
arr, mapping, mask_function=lambda arr, target: arr == target
):
res = arr.copy()
for old_value, new_value in mapping.items():
mask = mask_function(arr, old_value)
res[mask] = new_value
return res
It will work on simpler cases:
import numpy as np
arr = np.array([0, 1, 2, 3, 4, 5])
mapping = {1: 10, 3: 30, 5: 50}
res = ndarray_apply_mapping(arr, mapping)
assert np.all(res == [0, 10, 2, 30, 4, 50])
But also on more complicated cases as yours.
Let's say you have an array with a limited set of RGB values (or cluster labels resulting from k-means, or whatever):
import numpy as np
H, W, C = 8, 16, 3
vmin, vmax = 0, 255
num_values = 10
values = np.random.randint(vmin, vmax, size=(num_values, C))
values_rnd_idxs = np.random.randint(0, num_values, size=(H, W))
arr = values[values_rnd_idxs]
assert arr.shape == (H, W, C)
And you have a mapping from some of those values to new values:
new_values = np.random.randint(vmin, vmax, size=(num_values // 3, C))
mapping = {tuple(old): tuple(new) for old, new in zip(values, new_values)}
You can use this mapping as follows:
res = ndarray_apply_mapping(
arr,
mapping,
mask_function=lambda arr, target: np.all(arr == target, axis=-1),
)
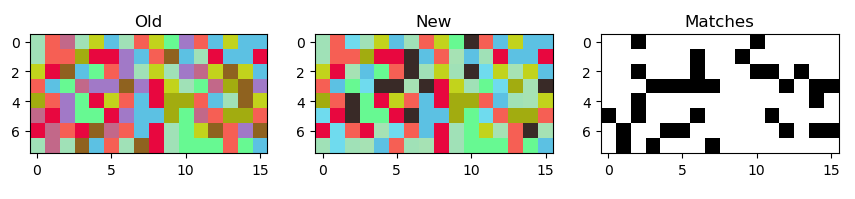
Plotting to see the result:
import matplotlib.pyplot as plt
fig, (ax_old, ax_new, ax_same) = plt.subplots(ncols=3)
ax_old.imshow(arr)
ax_new.imshow(res)
ax_same.imshow((res == arr).all(axis=-1), vmin=0, vmax=1, cmap="gray")
ax_old.set_title("Old")
ax_new.set_title("New")
ax_same.set_title("Matches")
plt.show()
CodePudding user response:
I should have caught it earlier just from your example data, but [30, 48, 29] does not exist in your example data:
[[ 29 48 30]
[ 29 48 30]
[ 29 48 30]
...
[ 29 48 30]
[ 29 48 30]
[ 29 48 30]]
...
[[111 137 58]
[111 137 58]
[111 137 58]
Somewhere along the line you inverted the color channels (RGB to BGR), and tried to compare a BGR color against RGB data. The match and replace line I suggested in the comments only needs a small modification if you want to keep the first variable in reverse order:
less_colors[(less_colors[:,:] == first[::-1]).all(axis=2)] = [0,0,0]