My Data:
library(lubridate)
library(tidyverse)
library(sf)
#test data columns
ID <- c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3)
start_date <- ymd_hms(c("2022-06-12 10:00:00", "2022-06-12 10:06:00", "2022-06-12 10:15:00", "2022-06-12 10:18:00",
"2022-06-12 10:01:00", "2022-06-12 10:05:00", "2022-06-12 10:08:00", "2022-06-12 10:15:00",
"2022-06-12 10:00:00", "2022-06-12 10:05:00", "2022-06-12 10:20:00"))
end_date <- ymd_hms(c("2022-06-12 10:05:00", "2022-06-12 10:10:00", "2022-06-12 10:25:00", "2022-06-12 10:20:00",
"2022-06-12 10:09:00", "2022-06-12 10:10:00", "2022-06-12 10:10:00", "2022-06-12 10:20:00",
"2022-06-12 10:00:00", "2022-06-12 10:13:00", "2022-06-12 10:26:00"))
start_lon <- c(1, 1, 2, 3, 1, 3, 2, 2, 3, 2, 4)
start_lat <- c(1, 3, 3, 2, 1, 2, 3, 3, 4, 2, 1)
end_lon <- c(2, 2, 2, 3, 1, 1, 1, 3, 2, 4, 1)
end_lat <- c(0, 3, 1, 1, 3, 1, 1, 1, 1, 2, 3)
#creating tibble
data <- tibble(ID, start_date, end_date, start_lon, start_lat, end_lon, end_lat)
# A tibble: 11 × 7
ID start_date end_date start_lon start_lat end_lon end_lat
<dbl> <dttm> <dttm> <dbl> <dbl> <dbl> <dbl>
1 1 2022-06-12 10:00:00 2022-06-12 10:05:00 1 1 2 0
2 1 2022-06-12 10:06:00 2022-06-12 10:10:00 1 3 2 3
3 1 2022-06-12 10:15:00 2022-06-12 10:25:00 2 3 2 1
4 1 2022-06-12 10:18:00 2022-06-12 10:20:00 3 2 3 1
5 2 2022-06-12 10:01:00 2022-06-12 10:09:00 1 1 1 3
6 2 2022-06-12 10:05:00 2022-06-12 10:10:00 3 2 1 1
7 2 2022-06-12 10:08:00 2022-06-12 10:10:00 2 3 1 1
8 2 2022-06-12 10:15:00 2022-06-12 10:20:00 2 3 3 1
9 3 2022-06-12 10:00:00 2022-06-12 10:00:00 3 4 2 1
10 3 2022-06-12 10:05:00 2022-06-12 10:13:00 2 2 4 2
11 3 2022-06-12 10:20:00 2022-06-12 10:26:00 4 1 1 3
What I want: Lines are to be created for each ID group. The lines are to be generated according to the time course of the points with the coorsinates lon and lat. There are always two points for each path (start and end) if the start point of a following path is before the end point of a previous path, this start point should be taken in between.
#Goal
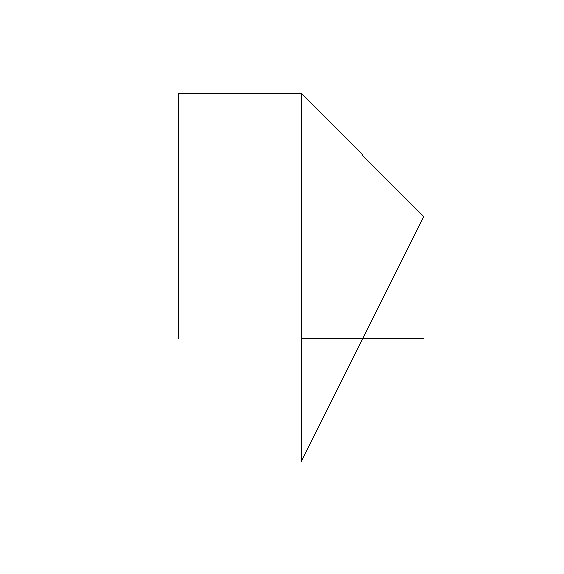
ID1_path_lon <- c(1, 2, 1, 2, 2, 3, 3, 2)
ID1_path_lat <- c(1, 0, 3, 3, 2, 2, 1, 1)
line_ID1 <- st_linestring(cbind(ID1_path_lon, ID1_path_lat))
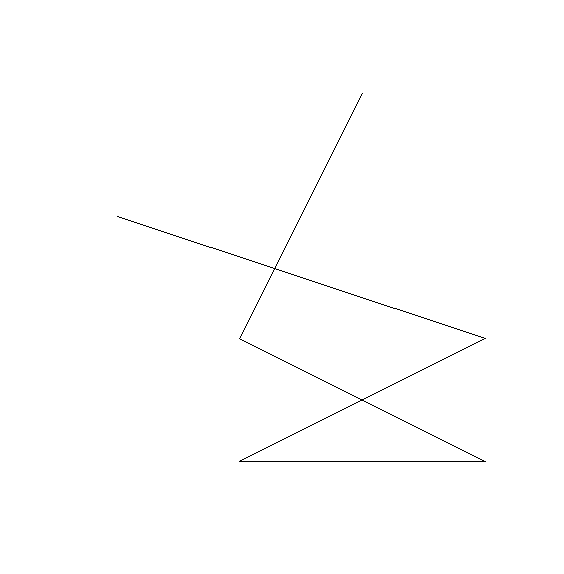
ID2_path_lon <- c(1, 3, 2, 1, 1, 1, 2, 3)
ID2_path_lat <- c(1, 2, 3, 3, 1, 1, 3, 1)
line_ID2 <- st_linestring(cbind(ID2_path_lon, ID2_path_lat))
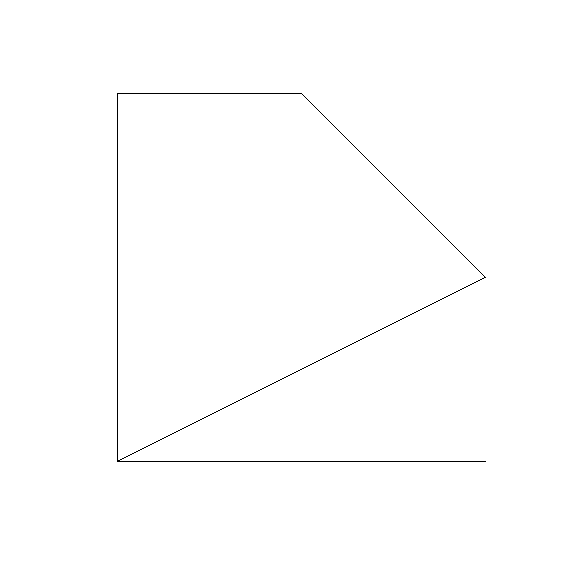
ID3_path_lon <- c(3, 2, 2, 4, 4, 1)
ID3_path_lat <- c(4, 1, 2, 2, 1, 1)
line_ID3 <- st_linestring(cbind(ID3_path_lon, ID3_path_lat))
plot(line_ID1, axes = TRUE)
plot(line_ID2, axes = TRUE)
plot(line_ID3, axes = TRUE)
My first attempts: My idea is to rebuild the data so that there is only one column for the date and two for the coordinates. This worked, but I get warnings, which makes me doubt if my solution is the best. Furthermore I have difficulties to sort by the date per group and then convert the whole thing as sf_linestring.
lines <- data %>% pivot_longer(cols = start_lon:end_lat) %>%
mutate(date = if_else(str_detect(name, "^start"), start_date, end_date)) %>%
mutate(latlon = if_else(str_detect(name, "lat$"), "lat", "lon")) %>%
select(ID, latlon, value, date) %>%
pivot_wider(names_from = latlon,
values_from = value) %>%
unnest(cols = lon:lat) %>%
arrange(ID, date) %>%
group_by(ID) %>%
st_linestring(rbind(lon, lat)) %>%
ungroup()
with the pivot_wider function I get a Warning message: Warnmeldung:
Values from `value` are not uniquely identified; output will contain list-cols.
* Use `values_fn = list` to suppress this warning.
* Use `values_fn = {summary_fun}` to summarise duplicates.
* Use the following dplyr code to identify duplicates.
{data} %>%
dplyr::group_by(ID, date, latlon) %>%
dplyr::summarise(n = dplyr::n(), .groups = "drop") %>%
dplyr::filter(n > 1L)
CodePudding user response:
If you do the pivot_longer() like this, this task becomes quite easy:
library(lubridate)
library(tidyverse)
library(sf)
data |>
pivot_longer(start_date:end_lat,
names_sep = "_",
names_to = c("time",".value")
) |>
arrange(ID, date) |>
group_by(ID) |>
summarize(line = st_sfc(st_linestring(cbind(lon,lat))))
#> # A tibble: 3 × 2
#> ID line
#> <dbl> <LINESTRING>
#> 1 1 (1 1, 2 0, 1 3, 2 3, 2 3, 3 2, 3 1, 2 1)
#> 2 2 (1 1, 3 2, 2 3, 1 3, 1 1, 1 1, 2 3, 3 1)
#> 3 3 (3 4, 2 1, 2 2, 4 2, 4 1, 1 3)