Brief summary
I have a matrix of values representing a topological surface. I'm trying to calculate the x values for an exact contour z for each column y.
In Depth
I'm fitting experimental data to a 3-dimensional non-linear decay model, z(x,y). Any code for my analysis needs to fit a number of different datasets. I've managed to parameterize the model successfully, and calculate z given each whole value of x and y. I then need to extract the values of specific contours. The range of each variable are x: (0 > x > 80); y: (0 > y > 100) ; z: (0 > z > 1).
require(pracma)
# Truncated matrix with demo data, normally created by fitting parameterized algorithm to 81x100 matrix
M <- matrix(c(0,1,1,1,1,1,1,1,1,
0,0.843,1,1,1,1,1,1,1,
0,0.484,0.907,1,1,1,1,1,1,
0,0.218,0.459,0.721,0.978,1,1,1,1,
0,0.082,0.185,0.313,0.461,0.621,0.781,0.925,1,
0,0.029,0.066,0.113,0.171,0.242,0.323,0.414,0.511,
0,0.010,0.022,0.038,0.058,0.082,0.112,0.146,0.186,
0,0.003,0.008,0.013,0.020,0.028,0.037,0.049,0.062,
0,0.001,0.003,0.004,0.007,0.009,0.012,0.016,0.021,
0,0.000,0.001,0.001,0.002,0.003,0.004,0.006,0.007,
0,0.000,0.000,0.001,0.001,0.001,0.001,0.002,0.002),ncol = 11)
# Extract Contours ----
X <- seq(from = 0, to = 80, by = 10) # normally by = 1 on full dataset
Y <- seq(from = 0, to = 100, by = 10)
z <- 0.8
FindZ <- function(x) interp2(X, Y, M, x, y)-z
x_out <- matrix()
for(i in 1:11){
y <- i*10 # adjusted for truncated dataset. Normally in increments of 1
x <- fzero(FindZ, mean(X))
x_out[i] <- x$x
print(x$x)
next}
I get the following error:
Error in if (fb == 0) return(list(x = b, fval = fb)) : missing value where TRUE/FALSE needed
I realize not all rows y will contain values exceeding the z contour I'm looking for at any given time. To circumvent I have attempted to use tryCatch() on my larger dataset . When I do this, I get nothing for say, up to loop iteration y[20], it runs successfully for approximately 15 iterations, then I get the same error for the balance. The problem being I have been getting this on rows where there are values >z, and the interpolation runs to an error. I have also switched the code, and sought solutions y for values of x and z, and achieved almost exactly the same result.
I don't want to subset the data as I don't want to lose the absolute x,y positions as they correspond to physical measurements. If there is no z value, I'd be expecting to place a 0 for the column using replace().
Ideally, I want a matrix of values of (x,y) that if I fed back into my original function, they would calculate to my value z. Any pointers would be great.
CodePudding user response:
I think you're trying to reinvent the wheel here. What you are attempting to do can be achieved efficiently using the isoband package, which can calculate the x, y co-ordinates of an exact z value given a matrix of z values with vectors of x, y co-ordinates:
result <- as.data.frame(isoband::isobands(Y, X, M, z, z)[[1]])
head(result)
#> x y id
#> 1 44.08998 80.00000 1
#> 2 44.08998 80.00000 1
#> 3 42.44618 70.00000 1
#> 4 40.00000 61.31944 1
#> 5 39.13242 60.00000 1
#> 6 35.27704 50.00000 1
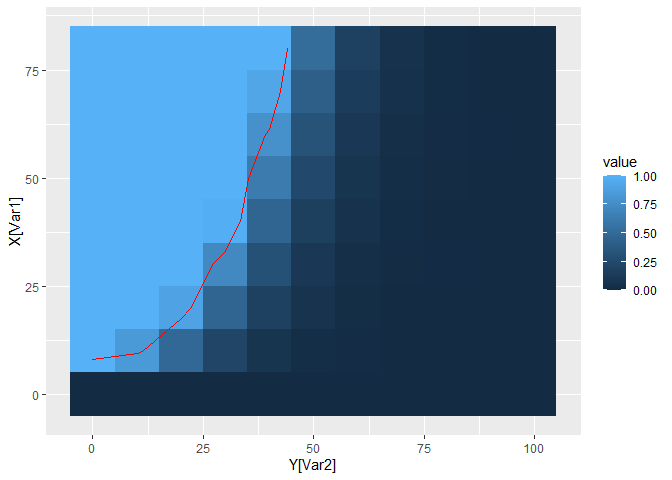
We can show this by plotting your grid with color representing the z values, and draw our result as a line.
library(ggplot2)
ggplot(reshape2::melt(M), aes(Y[Var2], X[Var1]))
geom_tile(aes(fill = value))
geom_path(data = result, aes(x , y), col = "red")
Created on 2022-08-08 by the reprex package (v2.0.1)
