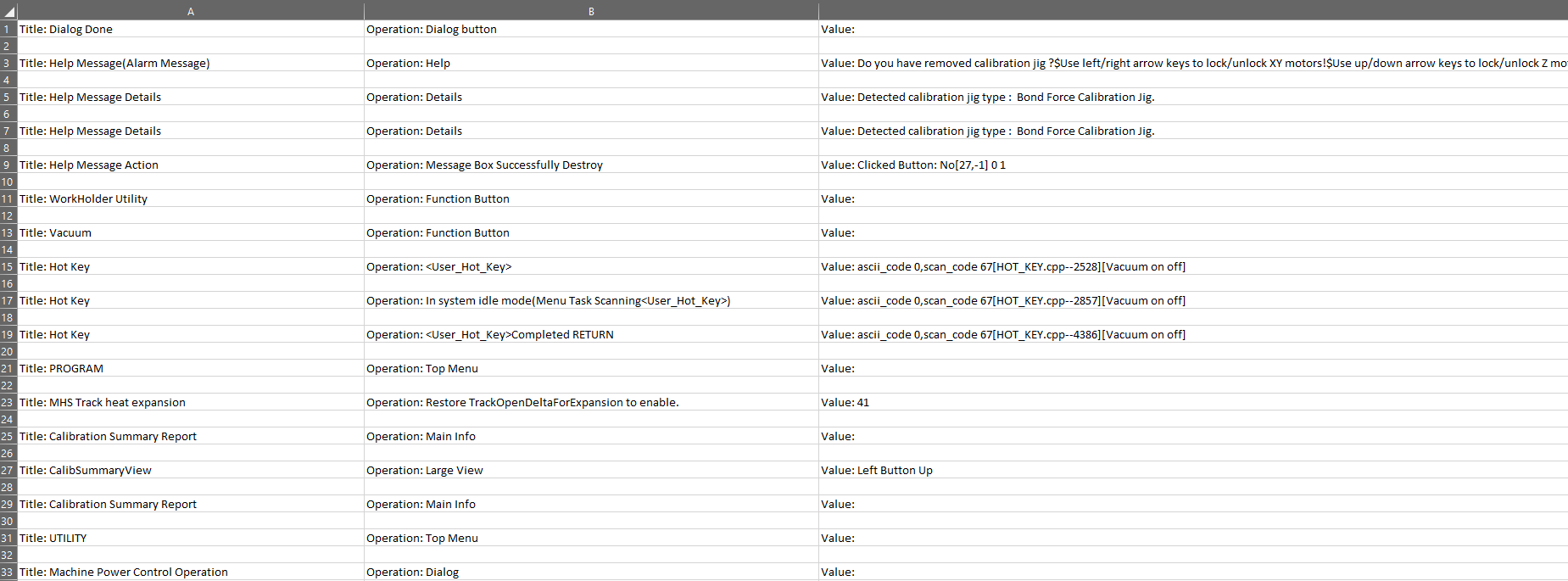
First, I already run through several question and answers here about my problem (Error tokenizing data) but I haven't got any working answer because I think my issue is not the same with the one I read.
I have a tab separated data (saved as .xls file) and be able to read thru pandas read_csv but encountered error on specific line during parsing. The data consist of ~58K rows with blank row every other row.
When reading the file through read_csv:
df = pd.read_csv('8176.xls', header=None, sep='\t', encoding='cp1252')
And this give an error: ParserError: Error tokenizing data. C error: Expected 12 fields in line 13245, saw 13
I read several thread that this could be an header error or a separator error.
I was able to locate the row/line which is encountering error through running my read_csv with nrows and it was in line 6623 of the data.
df = pd.read_csv('8176.xls', header=None, sep='\t', encoding='cp1252', nrows=6623)
Through readlines, I think this is the one causing the error:
There is unwanted tab delimiter (\t) inserted on this line:

The same issue when opening the file in excel, the data in that cell is splited and shifted by 1 column.
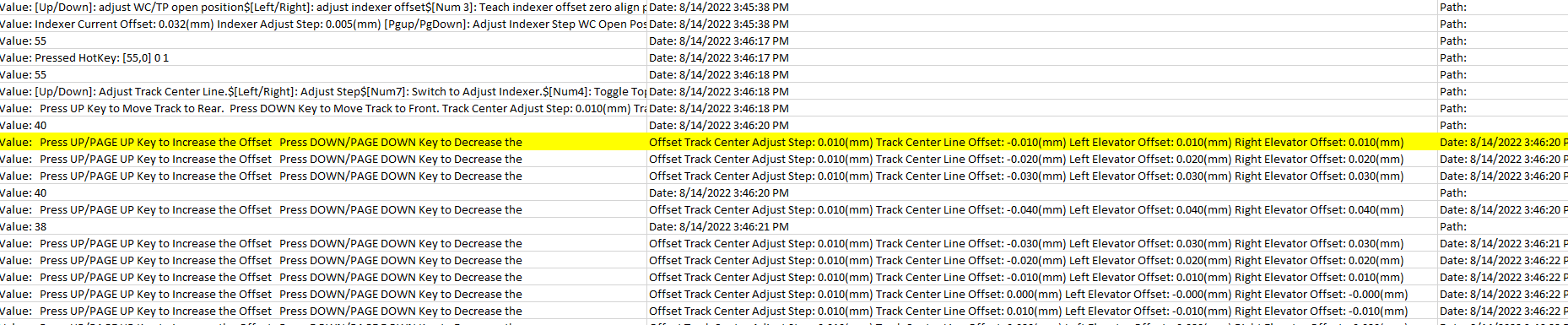
But when opening the file in notepad, it is only a space character not a Tab.

Is there any work around to eliminate the unwanted \t?
Or any encoding to read this as space only?
I already tried different encoding but also encountered errors.
Thank you so much.
CodePudding user response:
You can fix this programmatically during read_csv. Check this answer out:
Starting with pandas 1.4.0, read_csv() delivers capability that allows you to handle these situations in a more graceful and intelligent fashion by allowing a callable to be assigned to on_bad_lines=.
Pandas dataframe read_csv on bad data
CodePudding user response:
I don't know why you use read_csv to read the xsl file!? In my example, everything is well parsed by the read_excel function. True, I had to install engine “xlrd”. Excel file https://i.stack.imgur.com/fUyHG.png
df = pd.read_excel('8176.xls',header=None, na_filter=False)
print(df)
0 1 2
0 qqq rrr ttt
1
2 ff sss hh\th
3
4 aa sss hh\toff