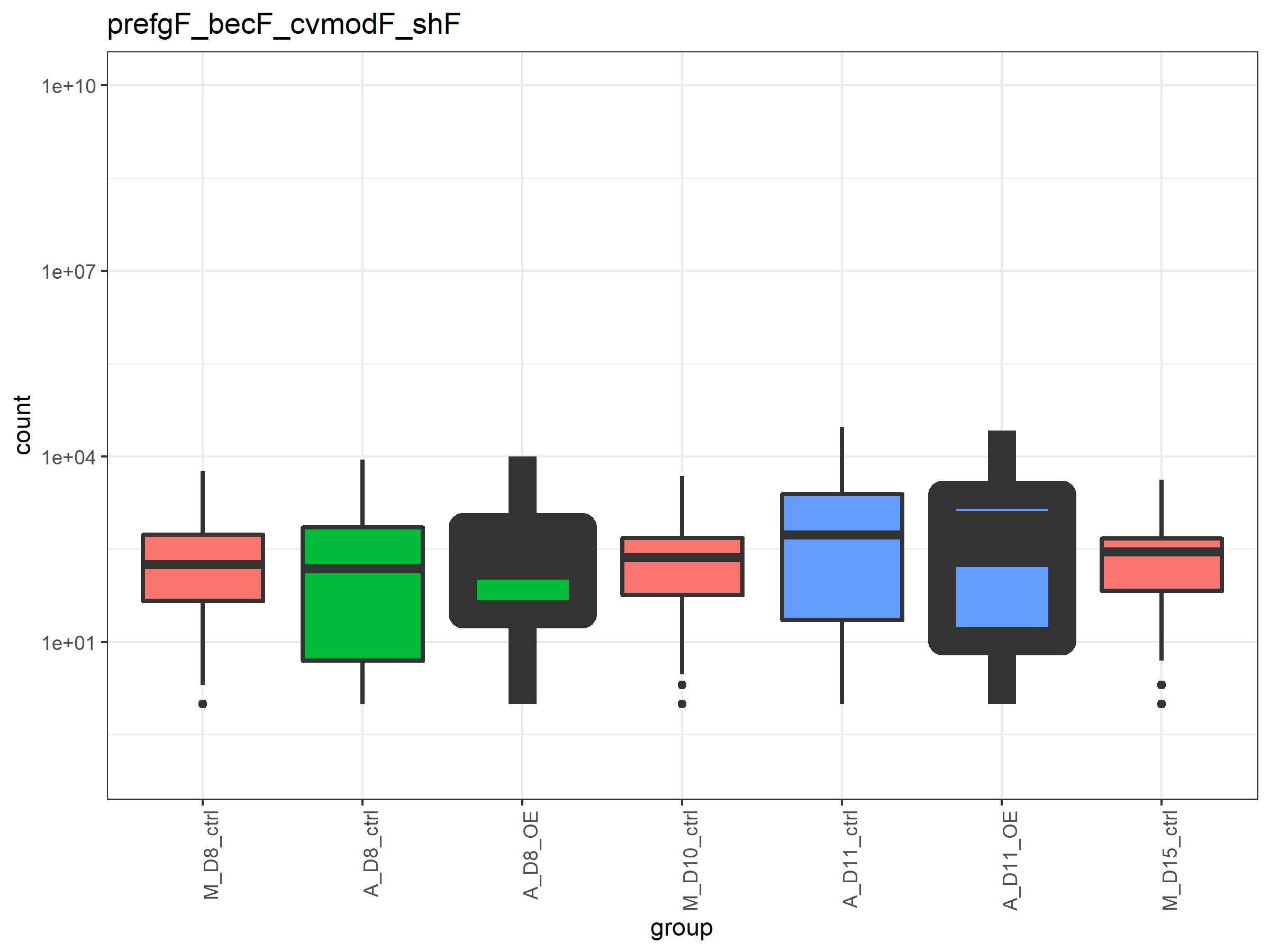
I am using the following code to generate a box plot:
df %>%
ggplot2::ggplot(ggplot2::aes(x = group, y = count, fill = batch))
ggplot2::geom_boxplot(ggplot2::aes(lwd = stroke))
ggplot2::scale_y_log10()
ggplot2::theme_bw()
ggplot2::theme(
axis.text.x = ggplot2::element_text(angle = 90, hjust = 1),
legend.position = "none"
)
ggplot2::labs(title = nm_dds)
I want the bold lines to be less thick, but adjusting the value of size_stroke has no effect. What am I doing wrong? Thanks.
In response to the comments, here is a reproducible example:
nm_dds <- "prefgF_becF_cvmodF_shF"
counts <- structure(c(0, 0, 2, 2906, 0, 0, 28, 0, 0, 0, 793, 5709, 4, 0,
1356, 90, 2, 0, 13, 268, 658, 669, 118, 666, 0, 880, 0, 46, 247,
100, 0, 0, 0, 0, 24, 671, 215, 6, 0, 72, 64, 544, 0, 227, 440,
2, 0, 0, 0, 366, 0, 0, 0, 2681, 0, 0, 47, 0, 0, 1, 707, 5439,
0, 0, 1246, 93, 0, 0, 23, 196, 547, 495, 113, 467, 0, 797, 0,
46, 226, 60, 0, 0, 0, 0, 14, 688, 173, 1, 0, 47, 52, 502, 0,
173, 390, 0, 0, 0, 0, 311, 0, 0, 0, 2284, 0, 0, 32, 0, 0, 0,
727, 5191, 1, 7, 1094, 100, 1, 0, 8, 179, 541, 499, 117, 448,
0, 691, 0, 49, 206, 78, 0, 0, 0, 0, 19, 585, 153, 6, 0, 87, 47,
510, 0, 169, 375, 3, 0, 0, 0, 281, 0, 0, 2, 1945, 0, 0, 54, 0,
0, 0, 633, 4791, 3, 2, 1340, 101, 0, 0, 7, 273, 519, 468, 112,
857, 0, 717, 0, 81, 302, 88, 0, 0, 0, 0, 20, 659, 156, 7, 0,
275, 66, 405, 0, 238, 330, 4, 0, 0, 0, 286, 0, 0, 0, 1912, 0,
0, 59, 0, 0, 0, 557, 4293, 5, 1, 1127, 104, 2, 0, 0, 252, 459,
481, 70, 724, 0, 642, 0, 77, 291, 81, 0, 0, 0, 0, 22, 551, 149,
4, 0, 241, 61, 393, 0, 185, 310, 2, 0, 0, 0, 285, 0, 0, 1, 1892,
0, 0, 39, 0, 0, 0, 497, 3656, 2, 3, 993, 94, 0, 0, 5, 217, 444,
461, 95, 550, 0, 557, 0, 89, 226, 86, 0, 0, 0, 0, 19, 503, 144,
0, 0, 304, 48, 380, 0, 188, 297, 4, 0, 0, 0, 256, 0, 0, 0, 1450,
0, 0, 45, 0, 0, 0, 537, 3359, 0, 2, 2182, 107, 2, 0, 1, 338,
472, 502, 82, 414, 0, 954, 0, 48, 335, 66, 0, 0, 0, 0, 40, 442,
139, 0, 0, 585, 86, 336, 0, 257, 290, 1, 0, 0, 1, 326, 0, 0,
0, 1598, 0, 0, 67, 0, 0, 0, 552, 3592, 0, 1, 1788, 92, 0, 0,
5, 371, 444, 407, 88, 459, 0, 852, 0, 48, 307, 69, 0, 0, 0, 0,
31, 422, 130, 1, 0, 731, 76, 338, 0, 264, 282, 1, 0, 0, 1, 284,
0, 0, 1, 1839, 0, 0, 62, 0, 0, 0, 576, 4176, 0, 0, 1743, 113,
1, 0, 0, 392, 483, 450, 87, 466, 0, 775, 0, 82, 357, 88, 0, 0,
0, 0, 34, 531, 157, 0, 0, 1055, 85, 313, 0, 240, 357, 2, 0, 0,
0, 322, 0, 0, 2, 2835, 0, 1, 92, 0, 0, 3, 1064, 7847, 1, 3, 2643,
113, 2, 0, 56, 356, 650, 918, 135, 1243, 0, 1466, 0, 105, 359,
81, 0, 0, 0, 0, 30, 905, 175, 3, 0, 102, 82, 675, 0, 344, 450,
1, 0, 0, 4, 512, 0, 0, 5, 3036, 0, 0, 94, 0, 0, 2, 1056, 8830,
3, 5, 2868, 108, 0, 0, 49, 365, 599, 900, 164, 1314, 0, 1483,
1, 154, 334, 100, 0, 0, 0, 0, 35, 996, 206, 3, 0, 165, 75, 710,
0, 363, 473, 3, 0, 0, 3, 541, 0, 0, 3, 2790, 0, 0, 83, 0, 0,
1, 982, 8658, 5, 4, 2709, 104, 1, 0, 43, 341, 639, 755, 127,
1272, 0, 1376, 0, 154, 331, 106, 0, 0, 0, 0, 24, 938, 227, 3,
0, 137, 62, 744, 1, 346, 485, 1, 0, 0, 2, 467, 0, 0, 4, 2870,
0, 0, 105, 0, 5, 2, 1576, 9938, 5, 4, 2515, 102, 1, 0, 72, 424,
690, 930, 136, 1261, 0, 746, 0, 317, 430, 93, 0, 0, 0, 0, 48,
993, 452, 18, 0, 44, 71, 517, 0, 414, 707, 0, 0, 0, 2, 509, 1,
0, 2, 2640, 0, 0, 86, 0, 4, 0, 1440, 8869, 1, 4, 2363, 96, 0,
0, 83, 375, 693, 1038, 114, 1158, 0, 740, 0, 286, 354, 99, 0,
1, 0, 0, 63, 1003, 388, 12, 0, 26, 65, 485, 0, 375, 650, 1, 0,
0, 5, 477, 0, 0, 1, 2816, 0, 0, 95, 0, 2, 2, 1485, 8370, 3, 6,
2391, 104, 1, 0, 61, 377, 747, 1040, 102, 1066, 0, 808, 0, 281,
352, 90, 0, 0, 0, 0, 50, 1017, 377, 17, 0, 30, 55, 514, 0, 399,
716, 0, 0, 0, 0, 468, 0, 0, 16, 7482, 0, 0, 323, 0, 0, 2, 3859,
30356, 9, 6, 11381, 512, 8, 0, 64, 1682, 3181, 3039, 664, 5347,
0, 8045, 0, 545, 2237, 542, 0, 1, 0, 0, 193, 3346, 772, 60, 0,
831, 533, 2611, 0, 1594, 2137, 3, 0, 0, 7, 2337, 0, 0, 4, 6041,
0, 2, 270, 0, 0, 1, 3379, 24967, 5, 5, 9458, 438, 10, 0, 37,
1171, 2601, 2768, 490, 4072, 0, 6498, 0, 300, 1849, 441, 0, 2,
0, 0, 161, 2708, 658, 32, 0, 582, 415, 2090, 0, 1358, 1786, 4,
0, 0, 10, 1845, 0, 0, 8, 6353, 0, 0, 314, 0, 0, 2, 3542, 28222,
3, 3, 9963, 443, 3, 0, 46, 1392, 2955, 2983, 578, 4262, 0, 8051,
0, 211, 2172, 554, 0, 4, 0, 0, 187, 2900, 673, 46, 0, 510, 514,
2332, 0, 1536, 1962, 14, 0, 0, 8, 2048, 1, 0, 13, 5209, 0, 2,
255, 0, 0, 1, 2982, 22262, 5, 11, 8089, 402, 5, 0, 45, 1131,
2392, 2527, 440, 3491, 0, 6795, 0, 143, 1722, 397, 0, 2, 0, 0,
158, 2409, 528, 39, 0, 459, 393, 1835, 0, 1221, 1708, 4, 0, 0,
1, 1660, 0, 0, 8, 3784, 0, 3, 298, 0, 2, 6, 3234, 26388, 187,
4, 8061, 433, 8, 0, 50, 1255, 3112, 2828, 454, 3934, 0, 2819,
2, 555, 1587, 471, 0, 4, 0, 0, 175, 2510, 1307, 30, 0, 218, 294,
1951, 0, 1443, 2315, 1, 0, 0, 6, 1617, 0, 0, 8, 4491, 0, 2, 262,
0, 3, 3, 3308, 19851, 160, 9, 9911, 332, 5, 0, 43, 1130, 2899,
3041, 483, 3595, 0, 3297, 3, 409, 1434, 593, 0, 5, 0, 0, 197,
2771, 1300, 52, 0, 209, 262, 1727, 0, 1466, 2285, 5, 0, 0, 7,
1956, 0, 0, 9, 6004, 0, 0, 328, 0, 4, 5, 3852, 24829, 139, 6,
10430, 454, 11, 0, 80, 1507, 3449, 3524, 514, 4097, 0, 3882,
2, 861, 1906, 566, 0, 0, 0, 0, 193, 3273, 1450, 40, 0, 470, 353,
2364, 0, 1627, 2628, 3, 0, 0, 4, 2066, 1, 0, 10, 4016, 0, 1,
217, 0, 3, 0, 2685, 16201, 137, 7, 7764, 315, 5, 0, 54, 989,
2412, 2611, 377, 2972, 0, 2820, 1, 615, 1224, 398, 0, 2, 0, 0,
146, 2421, 992, 42, 0, 400, 219, 1523, 0, 1131, 1909, 3, 0, 0,
5, 1409), .Dim = c(50L, 23L), .Dimnames = list(c("Pydc3", "Ceacam5",
"Mir93", "Idh1", "Gm10872", "C4bp-ps1", "Mrps33", "LOC432562",
"March1", "Rbm44", "Npc1", "Rpl8", "Nckap1l", "H2-Eb1", "Ghitm",
"Rabl5", "2700089I24Rik", "Vmn2r38", "Dpysl4", "Map2k3", "Tsc1",
"Kbtbd2", "Slc5a6", "Erbb2", "Olfr1104", "Tmem65", "Gpr142",
"Scube3", "B3gnt1", "A430033K04Rik", "Skint9", "4933411E08Rik",
"Olfr424", "Mir139", "Pcdhga4", "Cdc123", "Dpy19l3", "Ticam2",
"4930480K15Rik", "Igf1", "Slc41a3", "Uck2", "Guca1b", "Ppp6r2",
"Rab22a", "Csf2rb2", "Vmn1r72", "Vmn1r95", "Zap70", "Uqcc"),
c("IM_MR28_S28_L001", "IM_MR29_S29_L001", "IM_MR30_S30_L001",
"IM_MR31_S31_L001", "IM_MR32_S32_L001", "IM_MR33_S33_L001",
"IM_MR34_S34_L001", "IM_MR35_S35_L001", "IM_MR36_S36_L001",
"IM_AR_36", "IM_AR_37", "IM_AR_38", "IM_AR_39", "IM_AR_40",
"IM_AR_41", "IM_AR51_S1_L006", "IM_AR52_S2_L006", "IM_AR53_S3_L006",
"IM_AR54_S4_L006", "IM_AR55_S5_L006", "IM_AR56_S6_L006",
"IM_AR57_S7_L006", "IM_AR58_S8_L006")))
cd <- structure(list(batch = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L
), .Label = c("190701_K00242_0579_AH7GFLBBXY", "180810_K00242_0453_AHVJTJBBXX_IM_AR_RS21_180814_K00242_0456_AHW5LYBBXX_IM_AR_RS21_180817_K00242_0458_AHW57TBBXX_IM_AR_RS21",
"180406_K00242_0385_AHTCCHBBXX_IM_AR_RS8_180412_K00242_0388_AHT7NYBBXX_IM_AR_RS8",
"180814_K00242_0456_AHW5LYBBXX_IM_AR_RS21_180814_K00242_0456_AHW5LYBBXX_IM_AR_RS21_180817_K00242_0458_AHW57TBBXX_IM_AR_RS21",
"190322_K00242_0534_AHYLVWBBXX_IM_AM_RS8_dT"), class = "factor"),
time_label = c("D8", "D8", "D8", "D10", "D10", "D10", "D15",
"D15", "D15", "D8", "D8", "D8", "D8", "D8", "D8", "D11",
"D11", "D11", "D11", "D11", "D11", "D11", "D11"), time_value = c(8L,
8L, 8L, 10L, 10L, 10L, 15L, 15L, 15L, 8L, 8L, 8L, 8L, 8L,
8L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L), user = c("MR",
"MR", "MR", "MR", "MR", "MR", "MR", "MR", "MR", "AR", "AR",
"AR", "AR", "AR", "AR", "AR", "AR", "AR", "AR", "AR", "AR",
"AR", "AR"), treatment = structure(c(1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L), .Label = c("ctrl", "OE"), class = "factor"),
group = structure(c(10L, 10L, 10L, 11L, 11L, 11L, 12L, 12L,
12L, 17L, 17L, 17L, 18L, 18L, 18L, 19L, 19L, 19L, 19L, 20L,
20L, 20L, 20L), .Label = c("M_2i_ctrl", "M_D0_ctrl", "M_D1_ctrl",
"M_D2_ctrl", "M_D3_ctrl", "M_D4_ctrl", "M_D5_ctrl", "M_D6_ctrl",
"M_D7_ctrl", "M_D8_ctrl", "M_D10_ctrl", "M_D15_ctrl", "A_D6_OE",
"A_D6_ctrl", "A_D7_ctrl", "A_D7_OE", "A_D8_ctrl", "A_D8_OE",
"A_D11_ctrl", "A_D11_OE"), class = "factor"), group_pca_label = c("M_D8_ctrl",
NA, NA, "M_D10_ctrl", NA, NA, "M_D15_ctrl", NA, NA, "A_D8_ctrl",
NA, NA, "A_D8_OE", NA, NA, "A_D11_ctrl", NA, NA, NA, "A_D11_OE",
NA, NA, NA), condition = structure(c(10L, 10L, 10L, 11L,
11L, 11L, 12L, 12L, 12L, 10L, 10L, 10L, 15L, 15L, 15L, 16L,
16L, 16L, 16L, 17L, 17L, 17L, 17L), .Label = c("2i_ctrl",
"D0_ctrl", "D1_ctrl", "D2_ctrl", "D3_ctrl", "D4_ctrl", "D5_ctrl",
"D6_ctrl", "D7_ctrl", "D8_ctrl", "D10_ctrl", "D15_ctrl",
"D6_OE", "D7_OE", "D8_OE", "D11_ctrl", "D11_OE"), class = "factor"),
time = structure(c(10L, 10L, 10L, 11L, 11L, 11L, 13L, 13L,
13L, 10L, 10L, 10L, 10L, 10L, 10L, 12L, 12L, 12L, 12L, 12L,
12L, 12L, 12L), .Label = c("2i", "D0", "D1", "D2", "D3",
"D4", "D5", "D6", "D7", "D8", "D10", "D11", "D15"), class = "factor"),
sample = structure(1:23, .Label = c("IM_MR28_S28_L001", "IM_MR29_S29_L001",
"IM_MR30_S30_L001", "IM_MR31_S31_L001", "IM_MR32_S32_L001",
"IM_MR33_S33_L001", "IM_MR34_S34_L001", "IM_MR35_S35_L001",
"IM_MR36_S36_L001", "IM_AR_36", "IM_AR_37", "IM_AR_38", "IM_AR_39",
"IM_AR_40", "IM_AR_41", "IM_AR51_S1_L006", "IM_AR52_S2_L006",
"IM_AR53_S3_L006", "IM_AR54_S4_L006", "IM_AR55_S5_L006",
"IM_AR56_S6_L006", "IM_AR57_S7_L006", "IM_AR58_S8_L006"), class = "factor")), row.names = c("IM_MR28_S28_L001",
"IM_MR29_S29_L001", "IM_MR30_S30_L001", "IM_MR31_S31_L001", "IM_MR32_S32_L001",
"IM_MR33_S33_L001", "IM_MR34_S34_L001", "IM_MR35_S35_L001", "IM_MR36_S36_L001",
"IM_AR_36", "IM_AR_37", "IM_AR_38", "IM_AR_39", "IM_AR_40", "IM_AR_41",
"IM_AR51_S1_L006", "IM_AR52_S2_L006", "IM_AR53_S3_L006", "IM_AR54_S4_L006",
"IM_AR55_S5_L006", "IM_AR56_S6_L006", "IM_AR57_S7_L006", "IM_AR58_S8_L006"
), class = "data.frame")
long <- counts %>%
as.data.frame() %>%
tidyr::pivot_longer(
cols = everything(),
names_to = "sample",
values_to = "count"
) %>%
dplyr::left_join(cd, by = "sample")
long$sample <- factor(long$sample, levels = levels(cd$sample))
size_stroke <- 2
outline_treatment <- "OE"
long %>%
dplyr::arrange(time_value, treatment) %>%
dplyr::mutate(group = factor(group, levels = unique(group))) %>%
dplyr::mutate(stroke = dplyr::case_when(
treatment == outline_treatment ~ size_stroke,
TRUE ~ 0
)) %>%
ggplot2::ggplot(ggplot2::aes(x = group, y = count, fill = batch))
ggplot2::geom_boxplot(ggplot2::aes(lwd = stroke))
ggplot2::scale_y_log10(limits = c(0.1, 1E10))
ggplot2::theme_bw()
ggplot2::theme(
axis.text.x = ggplot2::element_text(angle = 90, hjust = 1),
legend.position = "none"
)
ggplot2::labs(title = nm_dds)
CodePudding user response:
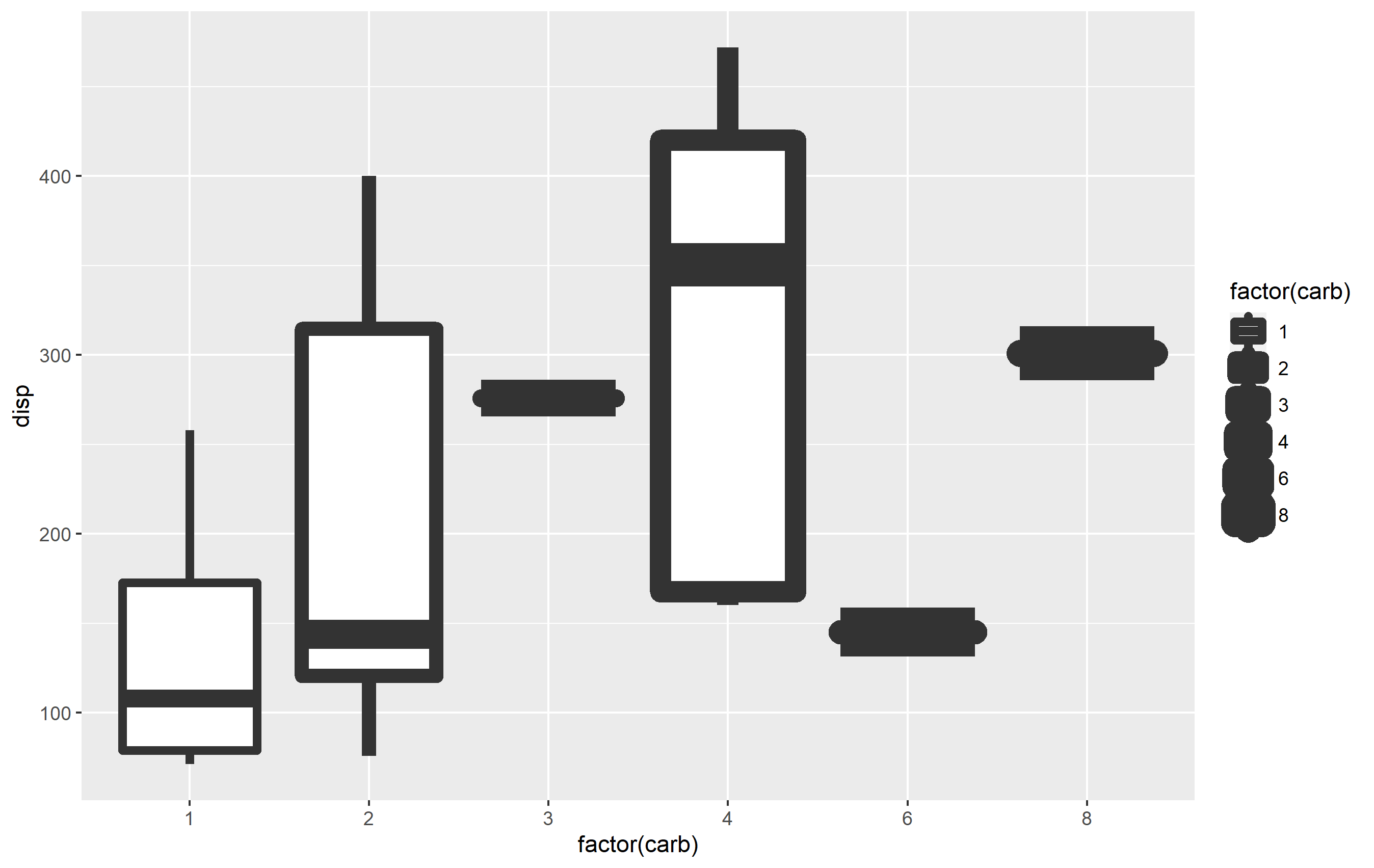
You can set the values for size via one of the scale_size_*() functions. Your reprex doesn't quite work without cd and a few other named objects in your environment, so I'm not sure what will work best for you; however, I can demonstrate an example of how this could work using mtcars.
library(ggplot2)
p <- mtcars %>%
ggplot(aes(x=factor(carb), y=disp))
geom_boxplot(aes(size=factor(carb)))
p
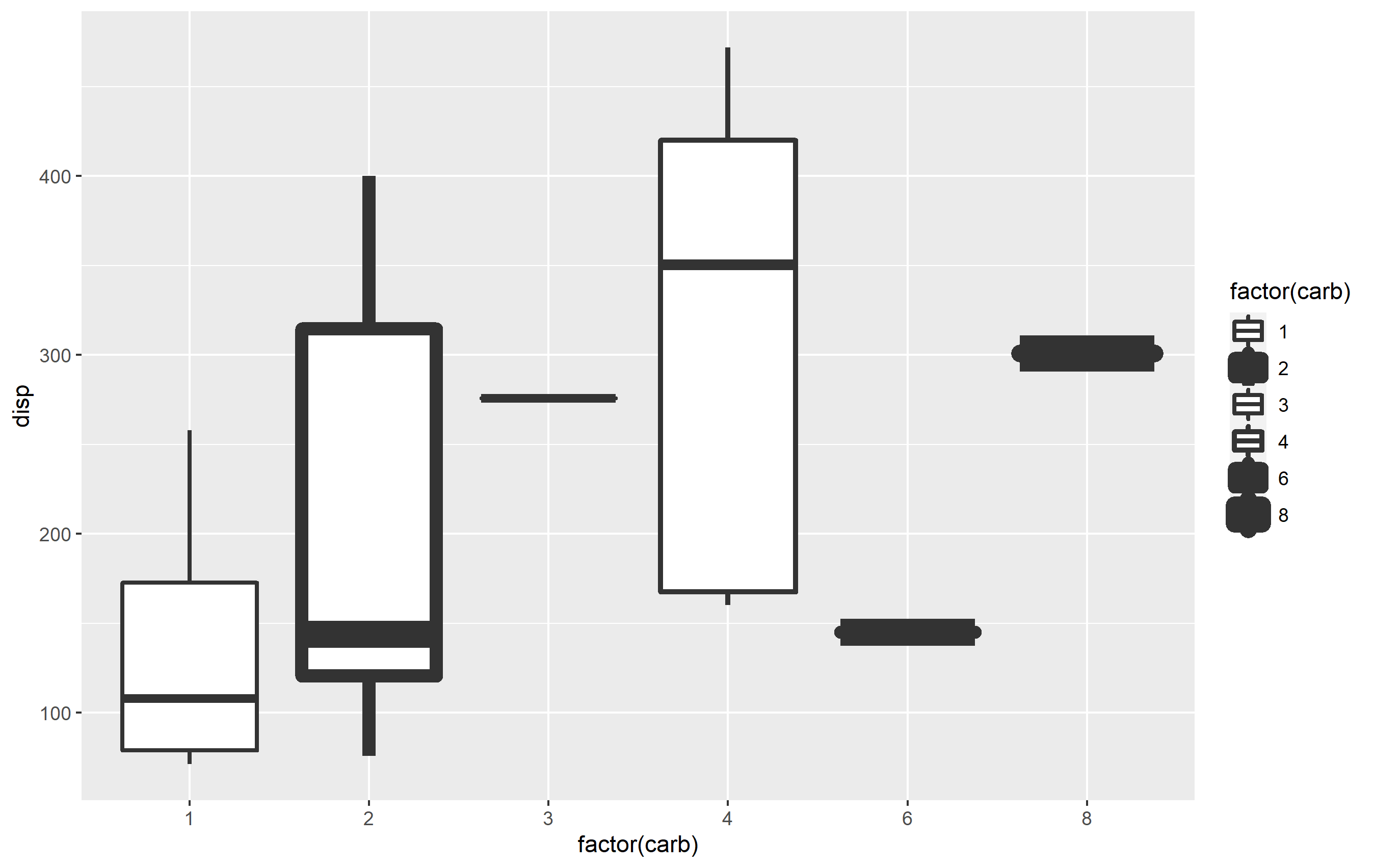
To set the sizes of each value manually, you can use scale_size_manual() and supply a values= argument as a vector which is then mapped to all levels of your factor. If you sent a named vector you can explictly assign the values to each level - otherwise the unnamed vector will map according to the level order.
p scale_size_manual(values = c(1,3,1,1.2,3,4))
Application to OP dataset
Thanks to the OP, we now have a dataset to work from :). If you apply the approach above directly to the OP's dataset, you encounter problems. I'll map size=stroke within geom_boxplot, just for the convenience of using the same aesthetic name (not lwd). It's helpful to separate out the data wrangling that happens before the plot code to ensure we understand what we're working with before you send it to plot:
d <- long %>%
dplyr::arrange(time_value, treatment) %>%
dplyr::mutate(group = factor(group, levels = unique(group))) %>%
dplyr::mutate(stroke = dplyr::case_when(
treatment == outline_treatment ~ size_stroke,
TRUE ~ 0
))
When you check the values in d$stroke using unique(d$stroke) we find only values of 0 and 2 exist. Theoretically, this means only two levels, but if you slap on scale_size_manual(values = c(0.5, 1.5)) to the code when using d, you get the following error message:
Error: Continuous value supplied to discrete scale
In addition: Warning messages:
1: Transformation introduced infinite values in continuous y-axis
2: Removed 405 rows containing non-finite values (stat_boxplot).
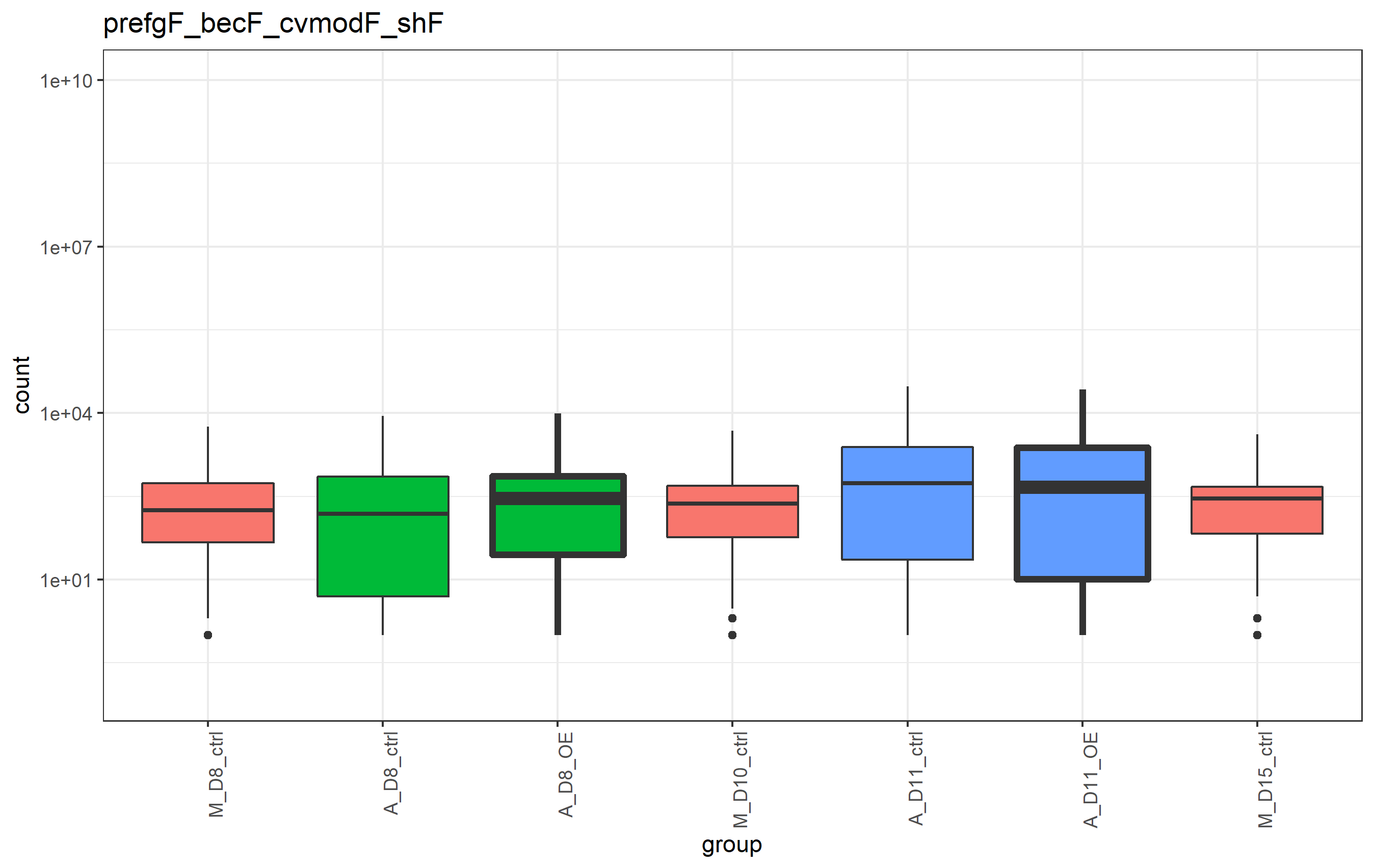
We can ignore the warnings (they deal with the y scale transformation and some NA values, but don't apply to the question at hand). Since d$stroke consists of only values of 0 or 2, it's a continuous column of values in the dataframe. Consequently, the size scale maps the value as if it was continuous. We could use scale_size_continuous, instead, but since I want to only have 2 discrete values, you can fix this by first converting d$stroke to a factor (forcing it to be discrete), then using scale_size_manual() at the end of your plot code. The full code to generate a fixed plot is as follows. Change the numbers in the values= argument for scale_size_manual() to change the look to the sizes you want:
# data wranglin'
d <- long %>%
dplyr::arrange(time_value, treatment) %>%
dplyr::mutate(group = factor(group, levels = unique(group))) %>%
dplyr::mutate(stroke = dplyr::case_when(
treatment == outline_treatment ~ size_stroke,
TRUE ~ 0
))
d$stroke <- factor(d$stroke) # need to convert to a factor if using scale_size_manual()
# plot code
d %>%
ggplot2::ggplot(ggplot2::aes(x = group, y = count, fill = batch))
ggplot2::geom_boxplot(ggplot2::aes(size = stroke))
ggplot2::scale_y_log10(limits = c(0.1, 1E10))
ggplot2::theme_bw()
ggplot2::theme(
axis.text.x = ggplot2::element_text(angle = 90, hjust = 1),
legend.position = 'none'
)
ggplot2::labs(title = nm_dds)
scale_size_manual(values=c(0.5, 1.5))