I am trying to plot a simple logistic regression in R.
I am following this tutorial to conduct the logistic regression and calculate a P-value (
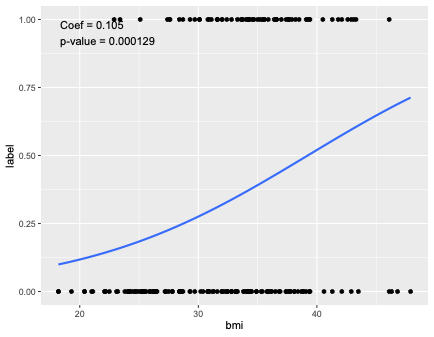
You have a glm, so glance() from tidy will not give you a p-value. Using an example:
library(ggplot2)
library(ggpmisc)
da = MASS::Pima.tr
da$label = as.numeric(da$type=="Yes")
model = glm(label ~ bmi,family=binomial,data=da)
summary(model)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -4.11156 0.92806 -4.430 9.41e-06 ***
bmi 0.10482 0.02738 3.829 0.000129 ***
You can see glance will not give you a p-value :
broom::glance(model)
# A tibble: 1 x 8
null.deviance df.null logLik AIC BIC deviance df.residual nobs
<dbl> <int> <dbl> <dbl> <dbl> <dbl> <int> <int>
1 256. 199 -120. 244. 251. 240. 198 200
You need to use tidy() and as @JonSpring mentioned in the comment, provide the formula, so something like this:
ggplot(da,aes(x = bmi,y = label)) geom_point()
stat_smooth(method="glm",se=FALSE,method.args = list(family=binomial))
stat_fit_tidy(method = "glm",method.args = list(family=binomial,formula=y~x),
mapping = aes(label = sprintf("Coef = %.3g\np-value = %.3g",
after_stat(x_estimate),after_stat(x_p.value))))
CodePudding user response:
Thank you for all the help, but unfortunately nothing automated worked, so I came up with this instead
require(cowplot)
require(ggplot2)
library(ggpmisc)
library(rms)
dataset=read.table('data.txt', header=TRUE)
model <- glm(variable ~ ancestry, data=dataset, family=binomial)
summary(model)
M1 <- glm(variable ~ ancestry, dataset, family = binomial)
M1
M1$null.deviance
M1$deviance
modelChi <- M1$null.deviance - M1$deviance
pseudo.R2 <- modelChi / M1$null.deviance
pseudo.R2
test <-lrm(variable ~ ancestry, dataset)
Chidf <- M1$df.null - M1$df.residual
chisq.prob <- 1 - pchisq(modelChi, Chidf)
chisq.prob
#plot logistic regression curve
all_variable <- ggplot(dataset, aes(x=ancestry, y=variable))
geom_point(alpha=.5, color=dataset$colorsite)
stat_smooth(method="glm", se=FALSE, method.args = list(family=binomial)) annotate("text", x=-Inf, y=Inf, hjust = 0, vjust = 2.5, label=paste("p-value: ",signif(chisq.prob, digits = 3),"\nR2: ",signif(pseudo.R2, digits = 3),sep="") )
ggtitle("Title not relevant to Stack Overflow")
ggsave("variable.pdf")