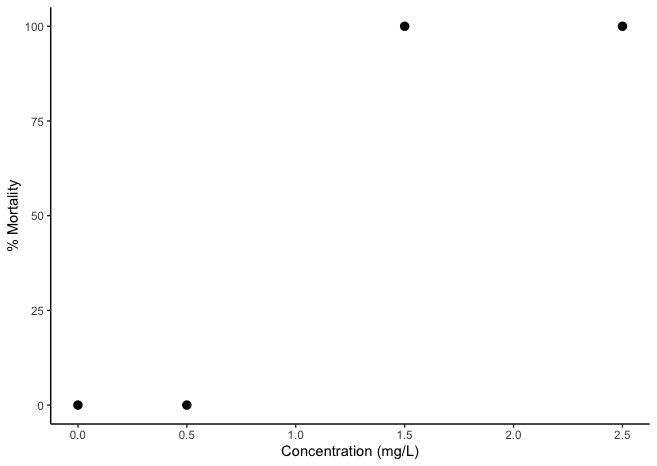
I need to graph a dose response curve for some bad data. How can I fit a line to this for an LC50?
lc50<- data.table(Concentration = c(0,0.5,1.5,2.5), Mortality= c(0,0,100,100))
ggplot(lc50,aes(Concentration, Mortality))
stat_summary(fun.data=mean_cl_normal)
scale_x_continuous(name="Concentration (mg/L)", limits=c(0, 2.5))
scale_y_continuous(name="% Mortality", limits=c(0, 100))
theme(panel.grid.major = element_blank(), panel.grid.minor =
element_blank(),panel.background =
element_blank(), axis.line =
element_line(colour = "black"))
CodePudding user response:
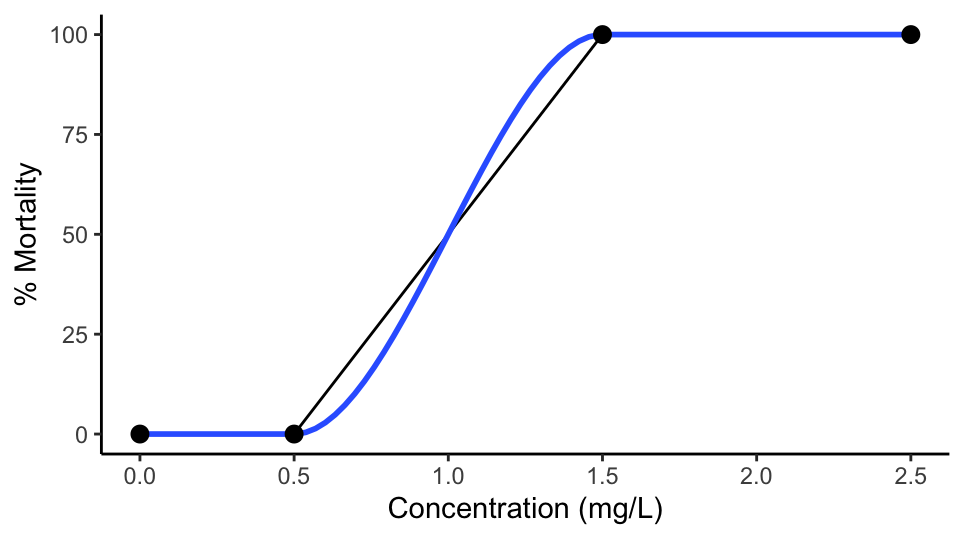
You can use this code:
lc50<- data.table(Concentration = c(0,0.5,1.5,2.5), Mortality= c(0,0,100,100))
ggplot(lc50,aes(Concentration, Mortality))
geom_line()
geom_smooth(method = "auto")
stat_summary(fun.data=mean_cl_normal)
scale_x_continuous(name="Concentration (mg/L)", limits=c(0, 2.5))
scale_y_continuous(name="% Mortality", limits=c(0, 100))
theme(panel.grid.major = element_blank(), panel.grid.minor =
element_blank(),panel.background =
element_blank(), axis.line =
element_line(colour = "black"))
Output:
CodePudding user response:
geom_line makes exact lines through all points, whereas geom_smooth(method="lm") will fit a single line with the smallest average distance to all points:
library(tidyverse)
library(data.table)
#>
#> Attaching package: 'data.table'
#> The following objects are masked from 'package:dplyr':
#>
#> between, first, last
#> The following object is masked from 'package:purrr':
#>
#> transpose
lc50 <- data.table(Concentration = c(0, 0.5, 1.5, 2.5), Mortality = c(0, 0, 100, 100))
ggplot(lc50, aes(Concentration, Mortality))
geom_line(color = "blue")
geom_smooth(method = "lm", color = "red")
stat_summary(fun.data = mean_cl_normal)
scale_x_continuous(name = "Concentration (mg/L)", limits = c(0, 2.5))
scale_y_continuous(name = "% Mortality", limits = c(0, 100))
theme(
panel.grid.major = element_blank(), panel.grid.minor =
element_blank(), panel.background =
element_blank(), axis.line =
element_line(colour = "black")
)
#> `geom_smooth()` using formula 'y ~ x'
#> Warning: Removed 14 rows containing missing values (geom_smooth).
#> Warning in max(ids, na.rm = TRUE): no non-missing arguments to max; returning
#> -Inf
#> Warning: Removed 4 rows containing missing values (geom_segment).
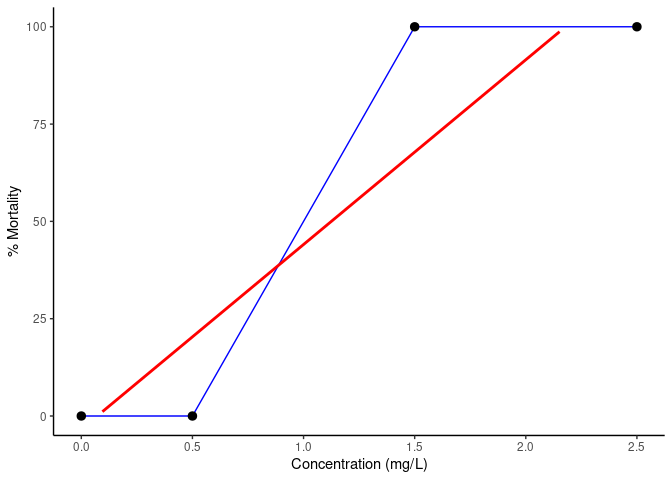
Created on 2022-03-03 by the reprex package (v2.0.0)