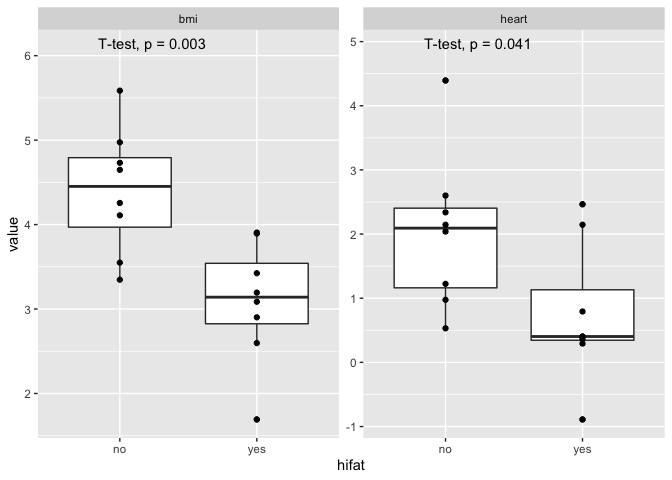
 I am looking at biological data of guinea pig with 2 treatment groups (hifat or no hifat diet) and when I facet_wrap the boxplots and add the stat_compare_means (t.test) function, the p-value is cut off. When I remove the scales="free", it still cuts off the p-value. I used the function to move the wording but since all graphs have different scales a fixed value for instance at y=1 would force all the axes to be the same. Would love any guidance.
I am looking at biological data of guinea pig with 2 treatment groups (hifat or no hifat diet) and when I facet_wrap the boxplots and add the stat_compare_means (t.test) function, the p-value is cut off. When I remove the scales="free", it still cuts off the p-value. I used the function to move the wording but since all graphs have different scales a fixed value for instance at y=1 would force all the axes to be the same. Would love any guidance.
library(tidyverse)
library(ggforce)
library(ggpubr)
ex <- data.frame(hifat=rep(c('yes','no'),each=8),
treat=rep(rep(c('bmi','heart'),4),each=4),
value=rnorm(32) rep(c(3,1,4,2),each=4))
ex %>%
ggplot(aes(x = hifat,
y = value))
geom_boxplot()
geom_point()
stat_compare_means(method = "t.test")
facet_wrap(~ treat, scales = "free")
CodePudding user response:
Edit
Thank you for editing your question to add an example dataset! Here is a potential solution:
library(tidyverse)
library(ggforce)
library(ggpubr)
ex <- data.frame(hifat=rep(c('yes','no'),each=8),
treat=rep(rep(c('bmi','heart'),4),each=4),
value=rnorm(32) rep(c(3,1,4,2),each=4))
ex %>%
ggplot(aes(x = hifat,
y = value))
geom_boxplot()
geom_point()
stat_compare_means(method = "t.test",
position = position_nudge(y = 0.5))
facet_wrap(~ treat, scales = "free")