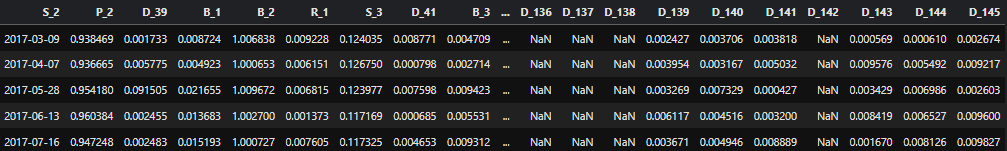
Given a large CSV file(large enough to exceed RAM), I want to read only specific columns following some patterns. The columns can be any of the following: S_0, S_1, ...D_1, D_2 etc. For example, a chunk from the data frame looks like this:
And the regex pattern would be for example anyu column that starts with S: S_\d.*.
Now, how do I apply this with pd.read_csv(/path/, __) to read the specific columns as mentioned?
CodePudding user response:
You can first read few rows and try DataFrame.filter to get possible columns
cols = pd.readcsv('path', nrows=10).filter(regex='S_\d*').columns
df = pd.readcsv('path', usecols=cols)
CodePudding user response:
Took the same approach(as of now) as mentioned in the comments. Here goes the detailed piece I used:
def extract_col_names(all_cols, pattern):
result = []
for col in all_cols:
if re.match(pattern, col):
result.append(col)
else:
continue
return result
extract_col_names(cols, pattern="S_\d ")
And it works! But without this work-around, say even loading the columns is heavy enough itself. So, does there exist any method to parse regex patterns at the time of reading CSVs? This still remains a question.
Thanks for the response :)