I need to contrast the my kl variable against the rtfpna variable across year and Contrast kl against ly, like this one:
I will be comparing labor productivity and labor intensity of output. y axis can be cumulative percent change. but i could not handle it. how can I plot it with ggplot?
my code is here so far:
data_long <- melt(brazil, id.vars = "year")
ggplot(data_long,
aes(x = year,
y = value,
col = variable))
geom_line()
but I can see only kl variable. I want to make Y axis percent change and other variables also should fit the graph. Can you help me please?
My data:
structure(list(year = c(1950, 1951, 1952, 1953, 1954, 1955, 1956,
1957, 1958, 1959, 1960, 1961, 1962, 1963, 1964, 1965, 1966, 1967,
1968, 1969, 1970, 1971, 1972, 1973, 1974, 1975, 1976, 1977, 1978,
1979, 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989,
1990, 1991, 1992, 1993, 1994, 1995, 1996, 1997, 1998, 1999, 2000,
2001, 2002, 2003, 2004, 2005, 2006, 2007, 2008, 2009, 2010, 2011,
2012, 2013, 2014, 2015, 2016, 2017, 2018, 2019), rtfpna = c(NA,
NA, NA, NA, 0.805425643920898, 0.819433450698853, 0.815486192703247,
0.850582480430603, 0.861791253089905, 0.875620603561401, 0.89727258682251,
0.9761683344841, 0.981472432613373, 1.00725114345551, 1.01003503799438,
1.03612756729126, 1.0297394990921, 1.04712808132172, 1.11110424995422,
1.10885286331177, 1.15965461730957, 1.2095251083374, 1.26581692695618,
1.29986226558685, 1.32612347602844, 1.31677484512329, 1.36736083030701,
1.31742632389069, 1.31152975559235, 1.36835289001465, 1.44977855682373,
1.3178858757019, 1.26234591007233, 1.19983005523682, 1.22336530685425,
1.24787294864655, 1.29066753387451, 1.28447079658508, 1.23783981800079,
1.24178087711334, 1.1565774679184, 1.14836263656616, 1.12104296684265,
1.14489555358887, 1.17703557014465, 1.18664479255676, 1.19274938106537,
1.18882584571838, 1.16212427616119, 1.11827838420868, 1.11366415023804,
1.10503828525543, 1.09412336349487, 1.08263611793518, 1.09423959255219,
1.0970253944397, 1.09963548183441, 1.13645040988922, 1.15163838863373,
1.12394869327545, 1.15129041671753, 1.13314604759216, 1.11766314506531,
1.11116170883179, 1.08192467689514, 1.03259932994843, 0.999256372451782,
1, 0.987977206707001, 0.97051203250885), kl = c(13384.1146710151,
13905.4317379675, 14543.3115078026, 14738.6663258408, 15117.962401934,
15419.2732598875, 15582.9626343674, 16085.4967356683, 16470.0290787947,
17186.6320696312, 17663.8533325066, 18305.7390664852, 18955.4212727199,
19315.4476234411, 19778.6872677136, 20348.143023004, 20964.6185147953,
21337.7172335079, 22004.2096224082, 23421.5943475649, 24886.7078912714,
25589.876623367, 26583.4606170811, 26239.5734578624, 28497.7343843658,
30784.0131388709, 32515.4201783311, 31395.0283935978, 32093.398044122,
34812.517528794, 38194.9191254873, 39465.210653577, 40021.7247790243,
41469.4399059998, 42145.3537526209, 41741.4306643921, 43918.1782180909,
45757.3252585928, 47590.1711487569, 50280.4650471358, 52346.3151598796,
56134.2221114178, 59940.6612822668, 63192.8950090397, 67099.8483065609,
71056.715493274, 76253.2274594481, 78581.3280983498, 81906.5419803117,
79683.3838413336, 78739.2803798882, 80408.9285072658, 79209.1317351792,
80408.8751312901, 79383.8858473091, 80864.9278521795, 84884.4177678533,
95936.2213458752, 111507.260345482, 132100.473013969, 149872.417184893,
159919.296530163, 158413.078594305, 155715.691961547, 153957.348437576,
150923.996107035, 149018.775987358, 148813.98192679, 147188.2646575,
145569.946224593), ly = c(0.000192081975169606, 0.000191676568301465,
0.000178150124840108, 0.000177261540902725, 0.000167979888410041,
0.000160545551853635, 0.00015930510632854, 0.000150759109766604,
0.00014667475219325, 0.000144362792203364, 0.000135573558523505,
0.000121862442568487, 0.00011598817899593, 0.000111137742890844,
0.000108993752154928, 0.000105077808249478, 0.000102753141141034,
9.71157787488221e-05, 9.02721802564341e-05, 9.11326210372743e-05,
8.21529112879634e-05, 7.74747056306122e-05, 7.29418232239867e-05,
7.2079948603802e-05, 6.78989033930588e-05, 6.68288701556521e-05,
6.32167379740309e-05, 6.77958027491811e-05, 6.72849300785159e-05,
6.18199857841665e-05, 6.10598843540433e-05, 6.13785967200655e-05,
6.322150328238e-05, 6.53099177091729e-05, 6.52259743232825e-05,
6.65807146866565e-05, 5.98779813321663e-05, 6.00058097132319e-05,
6.16151015847246e-05, 6.15075726816595e-05, 6.4564989654978e-05,
5.89804834703621e-05, 5.72009942535745e-05, 5.36571443523847e-05,
4.80415136646997e-05, 4.15694481109486e-05, 3.39854234488139e-05,
3.56472305678744e-05, 3.70922586244233e-05, 4.04059213895776e-05,
4.12019794519667e-05, 4.15933413003583e-05, 4.29868360536339e-05,
4.37847891035929e-05, 4.38000521738722e-05, 4.3492450941185e-05,
4.11081427705436e-05, 3.71200694703033e-05, 3.39197004604689e-05,
3.28454929411177e-05, 2.93904895197602e-05, 2.70128143683769e-05,
2.73584030910398e-05, 2.75700693997243e-05, 2.81523914533772e-05,
2.99867196457755e-05, 3.0914562397841e-05, 3.04839029945696e-05,
3.02719475151839e-05, 3.05049823911369e-05)), row.names = c(NA,
-70L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
The problem is that your three series have completely different scales. ly has values that are approximately 10,000 times smaller than rtfpna, and kl has values that are about 100,000 times greater than rtfpna. When you try to plot all the series together, the y axis will be large enough for you to see the values for all the series. That means it needs to range from 0 to 350,000. However, since the values for ly and rtfna are between 0 and 2, then they will sit along the very bottom pixel of your image. (if your plot is 1,000 pixels high, then anything with a value under 350 will be in the bottom pixel of the plot).
We can get all the series to appear on the plot if we divide kl by 100,000 and multiply ly by 10,000:
library(tidyverse)
p <- brazil %>%
mutate(kl = kl / 100000, ly = ly * 10000) %>%
pivot_longer(-year) %>%
ggplot(aes(year, value, color = name))
geom_line()
p
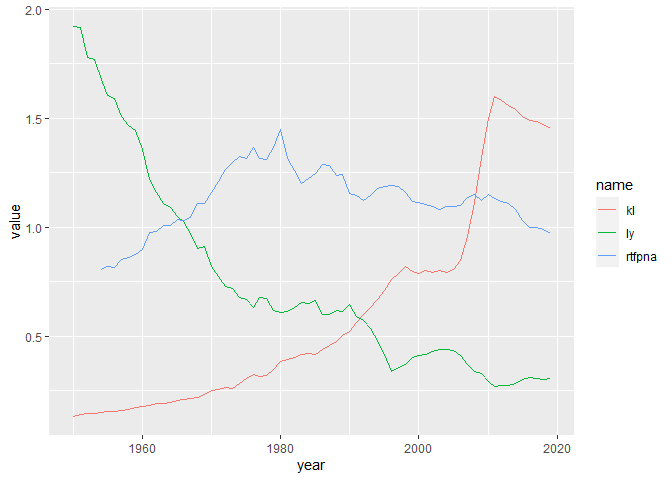
But now of course the problem is that the y axis scale does not give the correct readings for ly or kl. We can fix this to some extent by adding a second axis for kl
p scale_y_continuous(name = 'rtfpna',
sec.axis = sec_axis(~.x*200000, name = 'kl',
labels = scales::comma))
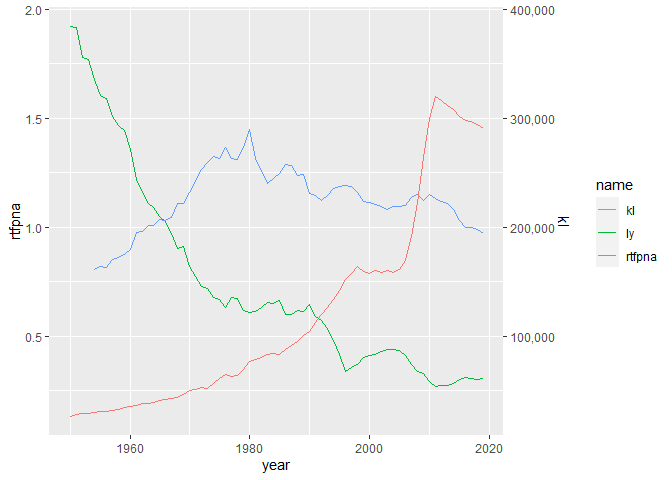
But the ly line still doesn't have a scale representing it.
It's generally considered poor practice to even have two different y axis scales on the same plot let alone three, and although there are hacks to achieve this, it's probably not worth doing.
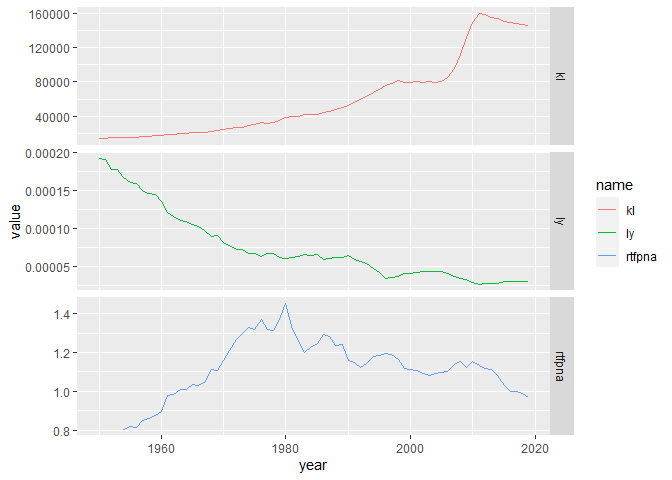
Your alternatives are to use facets:
brazil %>%
pivot_longer(-year) %>%
ggplot(aes(year, value, color = name))
geom_line()
facet_grid(name~., scales = 'free_y')
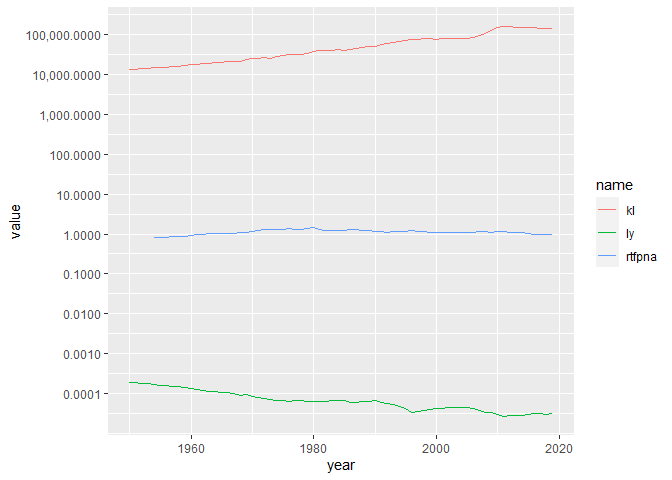
Or use a log axis to fit everything in:
brazil %>%
pivot_longer(-year) %>%
ggplot(aes(year, value, color = name))
geom_line()
scale_y_log10(breaks = 10^seq(-4, 5), labels = scales::comma)
Personally, I think facets are the way to go here.
Created on 2022-10-13 with reprex v2.0.2
