I am attempting to make a multi-panelled plot from three individual plots (see images).However, I am unable to rectify the bunched x-axis tick labels when the plots are in the multi-panel format. Following is the script for the individual plots and the multi-panel:
Individual Plot:
NewDat [[60]]
EstRes <- NewDat [[60]]
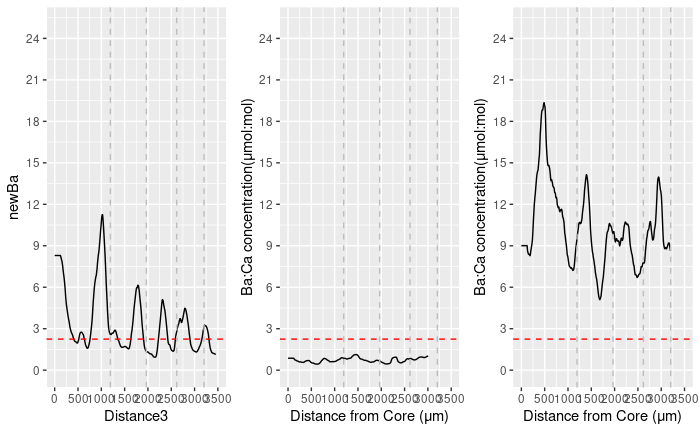
EstResPlt = ggplot(EstRes,aes(Distance3, `newBa`)) geom_line() scale_x_continuous(n.breaks = 10, limits = c(0, 3500)) scale_y_continuous(n.breaks = 10, limits = c(0,25)) xlab("Distance from Core (μm)") ylab("Ba:Ca concentration(μmol:mol)") geom_hline(yintercept=2.25, linetype="dashed", color = "red") geom_vline(xintercept = 1193.9, linetype="dashed", color = "grey") geom_vline(xintercept = 1965.5, linetype="dashed", color = "grey") geom_vline(xintercept = 2616.9, linetype="dashed", color = "grey") geom_vline(xintercept = 3202.8, linetype="dashed", color = "grey") geom_vline(xintercept = 3698.9, linetype="dashed", color = "grey")
EstResPlt
Multi-panel plot:
MultiP <- grid.arrange(MigrPlt,OcResPlt,EstResPlt, nrow =1)
I have attempted to include:
MultiP <- grid.arrange(MigrPlt,OcResPlt,EstResPlt, nrow =1)
theme(axis.text.x = element_text (angle = 45)) )
MultiP
but have only received errors. It's not necessary for all tick marks to be included. An initial, mid and end value is sufficient and therefore they would not need to all be included or angled. I'm just not sure how to do this. Assistance would be much appreciated.
CodePudding user response:
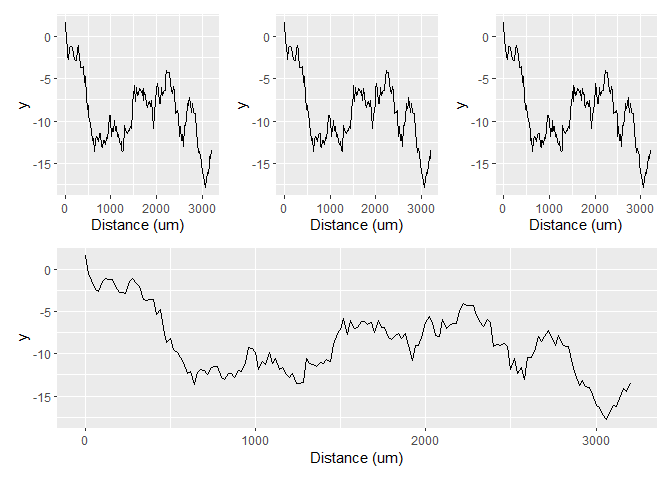
There are several options to resolve the crowded axes. Let's consider the following example which parallels your case. The default labelling strategy wouldn't overcrowd the x-axis.
library(ggplot2)
library(patchwork)
library(scales)
df <- data.frame(
x = seq(0, 3200, by = 20),
y = cumsum(rnorm(161))
)
p <- ggplot(df, aes(x, y))
geom_line()
(p p p) / p &
scale_x_continuous(
name = "Distance (um)"
)
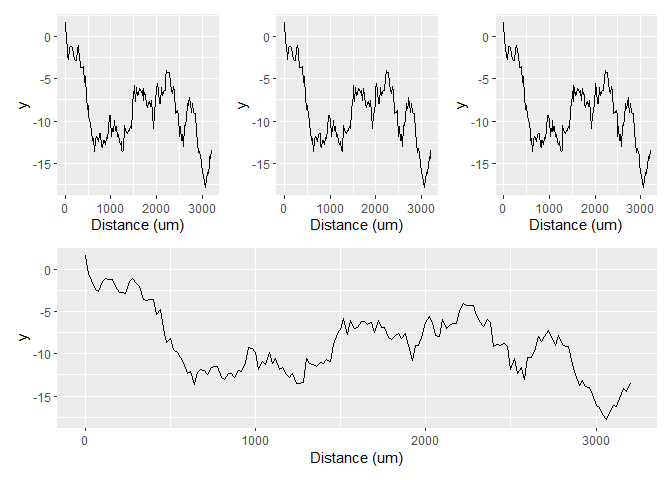
However, because you've given n.breaks = 10 to the scale, it becomes crowded. So a simple solution would just be to remove that.
(p p p) / p &
scale_x_continuous(
n.breaks = 10,
name = "Distance (um)"
)
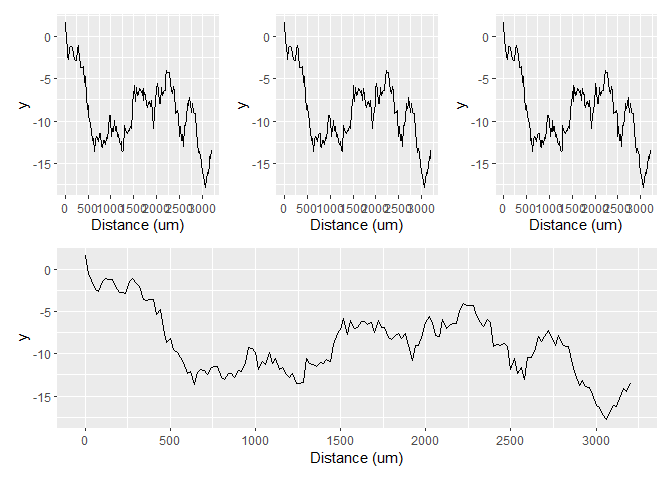
Alternatively, you could convert the micrometers to millimeters, which makes the labels less wide.
(p p p) / p &
scale_x_continuous(
n.breaks = 10,
labels = label_number(scale = 1e-3, accuracy = 0.1),
name = "Distance (mm)"
)
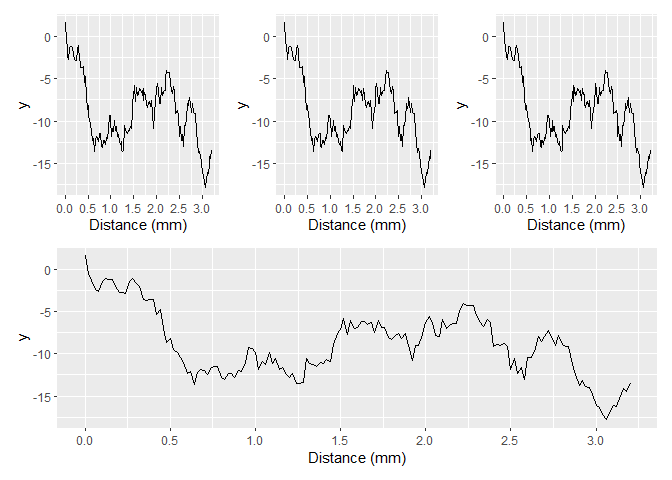
Yet another alternative is to put breaks only every n units, in the case below, a 1000. This happens to coincide with omitting n.breaks = 10 by chance.
(p p p) / p &
scale_x_continuous(
breaks = breaks_width(1000),
name = "Distance (um)"
)